MGMT promoter methylation in Peruvian patients with glioblastoma
Carolina Belmar-Lopez1, Carlos A Castaneda1,2, Miluska Castillo1, Pamela García-Corrochano3, Enrique Orrego3, Barbara Meléndez4, Sandro Casavilca5, Claudio Flores6 and Enrique Orrego3
1Department of Research, Instituto Nacional de Enfermedades Neoplasicas. Av Angamos Este 2520, Surquillo, Lima 15038, Peru
2Faculty of Medicine, Universidad Peruana San Juan Bautista. Av Jose Antonio Lavalle s/n, Chorrillos, Lima 15067, Peru
3Department of Neurosurgery, Instituto Nacional de Enfermedades Neoplasicas, Lima 15038, Peru
4Molecular Pathology Research Unit, Department of Pathology, Virgen de la Salud Hospital. Av De Barber s/n, Toledo 45005, Spain
5Department of Pathology, Instituto Nacional de Enfermedades Neoplasicas. Av Angamos Este 2520, Surquillo, Lima 15038, Peru
6Department of Research, Oncosalud. Av Guardia Civil 571, San Borja, Lima 15036, Peru
Correspondence to: Carlos A Castaneda. Email: carloscastanedaaltamirano@yahoo.com
Abstract
Purpose: O6-methylguanine–DNA methyltransferase (MGMT) promoter methylation predicts the outcome and response to alkylating chemotherapy in glioblastoma. The aim of this study is to evaluate the prevalence of MGMT methylation in Peruvian glioblastoma cases.
Patients and methods: We evaluated retrospectively 50 cases of resected glioblastoma during the period 2008–2013 at Instituto Nacional de Enfermedades Neoplasicas in Peru. Samples consisted of paraffin embedded and frozen tumour tissue. MGMT-promoter methylation status and the expression level of MGMT gene were evaluated by methylation-specific PCR and real-time PCR, respectively.
Results: Unmethylated, methylated and partially methylated statuses were found in 54%, 20% and 26% of paraffin-embedded samples, respectively. Methylation status was confirmed in the Virgen de la Salud Hospital and frozen samples. There was an association between the status of MGMT-promoter methylation and the level of gene expression (p = 0.001). Methylation was associated with increased progression-free survival (p = 0.002) and overall survival (OS) (p < 0.001).
Conclusion: MGMT-promoter methylation frequency in Peruvian glioblastoma is similar to that reported in other populations and the detection test has been standardised.
Keywords: MGMT, methylation, glioblastoma, Latin American
Copyright: © the authors; licensee ecancermedicalscience. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/3.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Published: 14/02/2018; Received: 07/09/2018
Background
Glioblastoma is the most common primary malignant brain tumour in adults and is invariably associated with an unfavourable prognostic. Patient survival rarely exceeds 12 months [1, 2]. Recent studies showed that the combined use of radiation and alkylating agents, such as temozolomide, increases the survival of patients with glioblastoma [3].
O6-methylguanine-DNA methyltransferase (MGMT) is a DNA repair enzyme that rapidly reverses alkylation of the O6 position of guanine and neutralises the cytotoxic effect of the mentioned alkylating agents [4]. The MGMT promoter contains a CpG island rich in Cytosine-Guanine (CG) dinucleotides or CpG sites and is positioned in the 5’ region of the same MGMT gene, located on the long arm of chromosome 10. Methylation of the CpG sites produces binding to specific proteins, and this complex causes altered chromatin structures and loss of transcription. Epigenetic silencing of the MGMT gene can be detected in around half of the glioblastoma cases and is associated with increased survival in patients receiving alkylating agents [5–11].
Stupp et al. evaluated the methylation status of MGMT-promoter in 206 glioblastomas through the specific PCR methylation test and found promoter methylation in 45%. It was associated with both longer overall survival (OS) and progression-free survival (PFS) in the treatment arm of concurrent radiation along with temozolomide (p < 0.007) [3, 6, 7].
The processes of methylation and other epigenetic changes are generated by the influence of environmental factors (e.g., food and pollution) and phenotypic characteristics (e.g., obesity) that vary according to the socio-cultural characteristics of populations [12].
The objective of the present study was the epidemiological evaluation of the methylation of the MGMT promoter in Peruvian patients with glioblastoma and the standardisation of the PCR technique in paraffin-embedded [formalin-fixed paraffin-embedded (FFPE)] samples from the Instituto Nacional de Enfermedades Neoplasicas (INEN) in Lima, Peru. This information will allow us, for the first time to our knowledge, to be aware of epidemiological information of epigenetic changes that behave as predictive biomarkers of response to treatment in the Peruvian population with implications for Public Health. The standardisation process includes four steps: (i) methylation status analysis of paired frozen and FFPE tumour samples (n = 7), and subsequent extension to 43 additional FFPE samples; (ii) validation test of methylation status in two evaluated FFPE samples at other institutions; (iii) MGMT gene expression analysis in frozen tumour (n = 7) and (iv) evaluation of the clinical behaviour of glioblastoma cases with methylated MGMT promoter.
Materials and methods
Study population
The study population consisted of 50 newly diagnosed glioblastoma cases resected during the period 2008–2013 at INEN with available FFPE samples. Seven cases had paired frozen tissue (ID: 21, 23, 24, 26, 27 and 40) stored at pathology department of our institute. Two FFPE samples (ID: 4 and 7) were also evaluated in the Molecular Pathology Research Unit of the Molecular Pathology Research Unit, Hospital Virgen de la Salud de Toledo, Spain (HVST).
Clinical and epidemiological information were obtained from the medical records and include: age, sex, initial Karnofsky performance status, tumour location, type of resection, adjuvant treatment, recurrence, recurrence date, follow-up date, and date of death.
Tumour area selection
Tumour samples were evaluated on haematoxylin–eosin slides for the identification of viable tumour areas (≥ 80%) with exclusion of necrotic, haemorrhagic or acute inflammatory reaction areas. A 0.6 cm cylinder was removed from the selected area and included in a new paraffin block.
Extraction of nucleic acids
For DNA extraction in frozen tissue, 15–20 mg from each sample was homogenised and purified using the GeneJET Genomic DNA Purification Kit (Thermo Scientific), following the manufacturer’s directions. For DNA and RNA extraction in FFPE samples, six slides (10-μm-thick sections) were processed and purified according to the commercial GeneJET FFPE DNA Purification kit (Thermo Scientific) and PureLink FFPE Total RNA Isolation Kit (Invitrogen, Life Technologies), repectively.
Quantification of nucleic acids
A fluorometric reading was performed to determine the concentrations of DNA and RNA obtained in the extractions. Measurements were carried out with Qubit (Invitrogen, Life Technologies) whose values are given in ng/μl. For this, a 1:200 working solution was prepared with the dsDNA/HS or RNA/HS fluorophore. For the quantification, 1 μl of DNA or RNA was added to the 199 μl working solution, and the samples were read.
DNA modification
Through treatment with bisulfite, unmethylated cytosine residues were converted to uracil. Two commercial kits were employed: (i) EpiJET Bisulfite Conversion Kit (Thermo Scientific), whereby 500 ng of the DNA was subjected to bisulfite, following the manufacturer’s directions. 120 μl of DNA-modifying reagent was added, and denaturation and subsequent conversion with bisulfite (98°C 10 minutes, 60°C 150 minutes) were performed. The DNA was then purified using the columns of the kit. (ii) EpiTect Bisulfite Kit (Qiagen), following the protocol, the reaction mixture containing 1 μg of the DNA, 85 μl of bisulfite mixture and 35 μl of protective DNA solution was prepared. The programme used for conversion with bisulfite was: 95°C 5 minutes; 60°C 25 minutes; 95°C 5 minutes; 60°C 85 minutes; 95°C 5 minutes and 60°C 175minutes. The DNA was then purified by using the kit columns.
Methylation-specific PCR (MSP)
After bisulfite treatment, 4 μl of the purified DNA was used to perform MSP. The PCRs were performed independently for methylated and unmethylated specific primers under variable conditions for annealing temperatures (55–66°C). All reactions were carried out using the optimised commercial mix Thermo Scientific Maxima Hot Start PCR Mix Master Green (Thermo Scientific). Each reaction mixture contains main reaction mixture, 400 mM sense primer, 400 mM antisense primer, 4 μl of bisulfite-treated DNA and nuclease-free water to make up to a final volume of 25 μl. Primer sequences for the methylated and unmethylated reactions have been reporter by Esteller et al. Sequence details for both forward and reverse primers are as follows: forward primer m_MGMT: 5′-TTTCGACGTTCTAGGTTTTCGC-3′; reverse primer m_MGMT: 5′-GCACTCTTCCGAAAACGAAACG-3′; forward primer um_MGMT: 5′-TTTGTGTTTTGATGTTTGTAGGTTTTTGT-3′; reverse primer um_MGMT: 5′-AACTCCACACTCTTCCAAAAACAAAACA-3’. In all reactions, a negative reaction control was added, in which the sample was replaced with water. Negative methylation controls were also added: peripheral blood normal lymphocytes and the LN18 cell line; and positive methylation controls: in vitro methylated normal blood lymphocytes in vitro with CpG Methyltransferase (M.SssI) (Thermo Scientific), and the LN18 cell line. The amplification was carried out in a Mastercycler Nexus Gradient (Eppendorf) with initial denaturing at 95°C for 5 minutes followed by 35 cycles of denaturing at 95°C for 50 seconds, annealing for 50 seconds at 55–66°C and extension for 50 seconds at 72°C, and then a final extension for 2 minutes at 72°C. The reaction products were analysed by electrophoresis on agarose gels (3%) and stained with a solution of SYBR Safe DNA gel stain (Invitrogen, Life Technologies). The unmethylated or methylated DNA amplicon size was 93bp and 81bp, respectively. The 50bp molecular weight marker (Invitrogen, Life Technologies) was used.
Reverse transcription from total RNA
SuperScript reverse transcriptase (Invitrogen, Life Technologies) was used to reverse transcription the RNA to cDNA. To do this, the following procedures were adopted: (i) 2 μg RNA, incubated with a 2U (1 U/μl) solution of DNase I (Invitrogen, Life Technologies) for 1 hour at 37°C; (ii) inactivation of DNase I with 2 μl of 25 mM ethylenediaminetetraacetic acid (EDTA) for 10 minutes at 65°C; (iii) incubation with 3 μl of random primers (Invitrogen, Life Technologies) and 1 μl of 10 mM dNTPs (Invitrogen, Life Technologies) for 5 minutes at 56°C, ending with 4°C; (iv) incubation with 5 μl of a ribonuclease inhibitor (40U) or RNaseOUT solution (Invitrogen, Life Technologies) for 2 minutes at room temperature; (v) incubating with 3 μl of a reverse transcriptase solution (200U) for 10 minutes at room temperature; (vi) incubating the mixture for 1 hour at 42°C and then for 15 minutes at 70°C. Control of the reaction was done in parallel without adding reverse transcriptase (called false reverse transcription). The resulting cDNA solution (50 ng/μl) was maintained at –20°C until use.
Real-time PCR (qPCR) using SYBR Green
The commercially available Fast Start Essential DNA Green Master (Roche) commercial blend was utilised for the reactions. Standard curve was performed to determinate the work concentration. Starting from a concentration of 100 ng/μl cDNA, three different sample dilutions (1/8, 1/16 and 1/32) were performed in duplicate. Each reaction mixture contained: 1X Fast Start Essential DNA Green Master, 400 mM sense primer, 400 nM antisense primer, 2 μl of corresponding cDNA dilution and nuclease free water to complete a final volume of 20 μl. The reactions were performed with the Light Cycler 96 System (Roche). Sequence details for both forward and reverse primers are as follows: forward primer 18S: 5′-CGGCTACCACATCCAAGGAA-3′; reverse primer 18S: 5′-GCTGGAATTACCGCGGCT-3′; forward primer MGMT: 5′-GCTGAATGCCTATTTCCACCA-3′; reverse primer MGMT: 5′-CACAACCTTCAGCAGCTTCCA-3′. Signal detection was performed at the end of the elongation step of the reaction. The conditions of the qPCR were: (i) 95°C 10 minutes and (ii) 95°C 15 seconds, 60°C 20 seconds, 72°C 20 seconds (45 cycles). The ratio of relative expression of genes to 18S was calculated using the ΔΔCq method. The Cq values were normalised to the values of the 18S housekeeping gene and the results were expressed as –ΔΔCq or Log2 (RQ), thus indicating the change in expression levels between the sample and the calibrator in the form of increments.
Data collection and statistical analysis
A database was made using the Microsoft Excel 2,000 programme (Microsoft Corporation, USA) with the variables collected without including the identity of the patients. Statistical analysis was performed using SPSS 12.0.0 software. (LEAD Technologies, Chicago, IL, USA). The analysis of the data included the study of the association of variables using the chi-square test and the t-Student test. The survival study was performed using the Kaplan–Meier method. The probability value p < 0.05 was accepted as significant.
Results
Clinical–pathological characteristics
Table 1 presents a summary of the clinical and pathological features of the 50 patients in the study, 40% were male and included 04 of ≤ 21 years old patients (8%). Most cases (58%) came in at least 90% Karnofsky at diagnosis. Histopathological diagnosis was performed under 2007 WHO classification. The temporal localisation was the most frequent (46%) and the median tumour size was 5 (1.5–7 cm). Most patients went to subtotal surgery (60%) and 40% to total surgery, and 74% received radiation, and 60% received chemotherapy, of which 83.33% received temozolomide.
DNA status in frozen and paraffin tissue samples
In order to evaluate the quality and degradation degree of the DNA obtained from the seven cases in which FFPE and frozen tumour tissue were available, the samples were quantified, analysed by amplification of the 18S ribosomal gene (18S rRNA) and visualised in 3% agarose gel (Figure 1). The results had shown a higher degree of degradation and loss of band intensity in the FFPE samples when compared with the same frozen sample.
Table 1. Clinical and pathological characteristics.
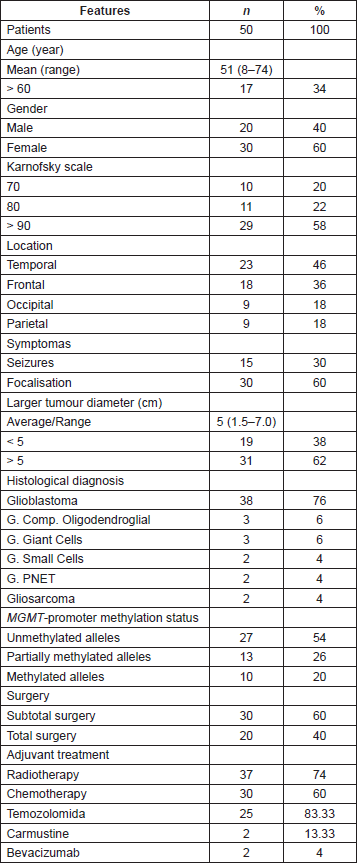
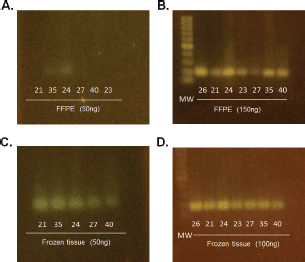
Figure 1. Determination of DNA status in paired paraffin and frozen tissue samples. Visualisation of amplification of the 18S ribosomal gene (18S rRNA) on 3% agarose gel. (A) and (C) Comparison of tumour stored in paraffin-embedded tissue versus frozen in 50ng. (B) and (D) Comparison of tumour stored in paraffin- embedded tissue versus frozen in 150 ng and 100ng, respectively.
MGMT-promoter methylation status
The methylation status of MGMT-promoter was determined in all tumours (n = 50) by MSP. Unmethylated (U) alleles were found in 54% (n = 27) of the tumours, methylation (M) of both alleles in 20% (n = 10) and methylation of only one allele (partial: P) in 26% (n = 13) of the tumours (Figure 2). We did not find differences for the methylation status according to age (p = 0.164), sex (p = 0.647) or tumour size (p = 0.876).
The results of methylation status in the HVST experiments were the same as those obtained in INEN, one case with methylated alleles (ID: 4) and another with partial methylation (ID: 7).
The results obtained for paired samples from FFPE and frozen tissue were similar, with partial methylation (ID: 26, 27, 35 and 40, n = 4/7) and the absence of methylation in both alleles (ID: 21, 23 and 24; n = 3/7). None of the tumour samples had both methylated alleles (n = 0/7).
Association between expression level of the MGMT gene and MGMT-promoter methylation status
The expression level of the MGMT gene was determined by real-time PCR (qPCR) in 32.7% of tumours (n = 17/50) (Figure 3). The remaining samples could not be evaluated due to poor quality and low concentration at obtained RNA. Expression levels were normalised to the 18s housekeeping gene. Values obtained from samples of healthy brain tissue were used as a calibrator. The results were expressed as -–ΔΔCq, as they evaluated the variation in the expression levels between the samples and the calibrator.
The association analysis between MGMT promoter methylation status and MGMT gene expression level showed significant differences (p = 0.001). The lowest expression levels were found in samples with both methylated alleles (n = 3) and had an average of 7.66 ± 0.42 times less than the control. The highest expression levels were observed in the samples with unmethylated alleles (n = 7) and had an average of 0.28 ± 0.38 times less than the control. Expression levels in samples with partial methylation (n = 7) (one methylated and one unmethylated allele) averaged 3.18 ± 0.40 times less than the control.
Association between MGMT-promoter methylation status and PFS
The results obtained together with the analysis of clinical and epidemiological information showed a significant association between PFS and all the MGMT-promoter methylation status (p = 0.002), sex (p = 0.029) and degree of resection (p = 0.001) (Figure 4). However, no significant association was found between PFS and any age (p = 0.527), Karnofsky (p = 0.535) and tumour size (p = 0.5) variables. According to the Cox regression model, a significant association remained for the presence of methylation (p = 0.01) and degree of resection (p = 0.004).
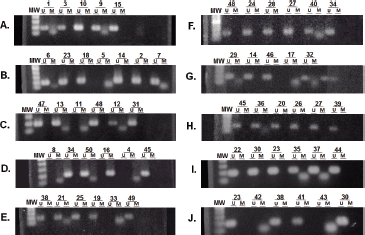
Figure 2. Determination of the methylation status of the MGMT promoter. Samples were analysed by MSP. The amplification of the PCR was for unmethylated alleles (U) and methylated alleles (M). The results were visualised on 3% agarose gel. The samples analysed were: (A) 1, 3, 10, 9 and 15. (B) 6, 23, 18, 5, 14, 2 and 7. (C) 47, 13, 11, 48, 12 and 31. (D) 8, 34, 50, 16, 4 and 45. (E) 38, 21, 25, 19, 33 and 49. (F) 48, 24, 28, 27, 40 and 34. (G) 29, 14, 46, 17 and 32. (H) 45, 36, 20, 26, 27 and 39. (I) 22, 30, 23, 35, 37 and 44. (J) 23, 42, 38, 41, 43 and 30.
Figure 3. Evaluation of gene expression level of MGMT by qPCR. Expression levels of MGMT gene were expressed as increments –ΔΔCq and indicated the change in expression levels between tumour samples and the brain tissue calibrator in the form of increments. (A) Expression level of MGMT gene for samples with both methylated alleles (M) and with partial methylation (one methylated and one unmethylated allele) (P). (B) Expression level of MGMT gene for samples with both unmethylated alleles (U).
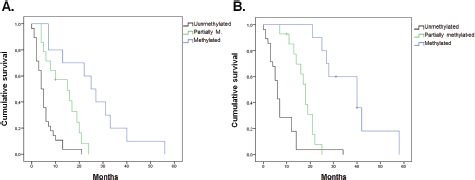
Figure 4. Estimated curve of PFS regarding MGMT-promoter methylation status. (A) Estimated curve of OS regarding MGMT-promoter methylation status. (B)
Association between MGMT-promoter methylation status and the global survival
The results showed a significant association between OS and both MGMT-promoter methylation status (p < 0.001) and degree of resection (p < 0.001) (Figure 4). No significant association was found for age (p = 0.198), sex (p = 0.085), Karnofsky (p = 0.318) nor tumour size (p = 0.863). According to the Cox regression model, a significant association remained methylation status (p < 0.001) and degree of resection (p < 0.001).
Discussion
Establishing and implementing predictive markers of prognosis and response to treatment in clinical practice allow a better patient selection for treatment. The review of the fourth edition of the recently published 2016 WHO Classification has begun to introduce molecular markers including methylation status of MGMT promoter; however, most considered information is still limited to traditional anatomo-pathological criteria [13].
Cytosine methylation in different areas of the genome, as in the case of the MGMT promoter, has specific patterns associated with race and environmental factors such as diet quality [14, 15] and its effect on the behaviour of the neoplasia may differ according to race [16]. Some studies found that glioblastoma has a higher prevalence in non-Hispanics than in Hispanic population [17, 18]. The present study describes, for the first time to our knowledge, a rate of MGMT-promoter methylation status in Peruvian cases of glioblastoma (46%) that is in the range described for Caucasian race (36–50%) [19–24].
To evaluate the quality of our samples, we compared the quality and degree of degradation of DNA obtained from FFPE and from frozen samples. The evaluation revealed a higher degree of degradation and loss of band intensity in the FFPE samples. Similarly, Sanchez-Navarro et al evaluated the quantitative data correlation between fresh and FFPE tissue samples. They found that the RNA extracted from FFPE samples had shown a variation in the basal values of Cq that can be explained by extensive degradation. However, adequate normalisation may compensate the effects of RNA degradation on the measurement of gene expression. They also found that the correlation among normalised expression values is better for genes of moderate-high expression [25, 26].
The analysis of two paraffin samples was repeated in other institution and confirmed our results. A pilot study comparing protocols for determination of the MGMT promoter status in 23 centres in Germany, Austria and the Netherlands found a good concordance rate in the group with methylation of both alleles, whereas differed according to the laboratory in the group with partial methylation [27]. The conference on the European consensus for the measurement of external quality in molecular pathology carried out in 2012 recommended the implementation of certification programs that include standardised inter-laboratory comparison at international level [28].
We evaluated the relationship between promoter methylation status and MGMT gene expression in 17 cases, and found an adequate correlation for the group of cases with unmethylated alleles and for the methylated allele group (p = 0.001). Similarly, Bady et al compared the promoter status and MGMT gene expression in the same samples and found an adequate correlation between both values and between these values and survival [29]. MSP only indicates whether or not the cytosine residues are methylated or not in the MGMT promoter, but not, if there is loss of expression of the MGMT gene. Therefore, direct analysis of the expression of MGMT allows corroborating that there are no alternative routes or modifications [24].
Testing methylation of MGMT promoter in glioblastoma has been implemented in centres of reference in cancer worldwide and our results confirm the prognostic value of the methylation status in our population [5, 19, 21, 22, 24]. Thus, we can state that the methodology used in our study to detect epigenetic changes in tumours can be reliably implemented in the routine practice of glioblastoma patients in our cancer institute, and our analysis could be replicated and implemented in other Latin American centres. Incorporation of this technique will allow to select those cases with greater benefit to alkylating chemotherapy and to classify the Latin American population under molecular classification.
The strength of our study is that it used the PCR methodology which is the test of choice for evaluating methylation because of its simplicity and easy access which allows its implementation in Latin American countries [19–24]. Some recent studies have suggested that a quantitative analysis of the MGMT-promoter methylation through sequencing technology correlates better with survival, however, it is a more expensive methodology [30].
A weakness of our series is the small size sample that needs to be confirmed in other Latin American series.
Conclusion
The rate of MGMT-promoter methylation in Peruvian glioblastoma cases is similar to that described in other races and PCR for its detection in FFPE of glioblastoma samples is an accurate methodology to implement in Latin American countries.
Conflict of interests
The authors declare that they have no conflict of interests.
Institutional review
The conduct of this survey was approved by The Institutional Review Board of INEN (#051-2016-CRP-DI-DICON/INEN). Since the study was based on a secondary source and there was no contact with the patients, no informed consent was applied; however, the identity and personal data of patients’ medical records were protected at all times.
Authors’ contributions
Belmar-Lopez C, Castaneda CA, Castillo M, Garcia-Corrochano P and Orrego E contributed to the conception and design of the study and performed data analysis and interpretation; Casavilca S, Barbara M and Claudio F performed the data acquisition, as well as providing administrative, technical, and material support; all authors drafted the article and made critical revisions related to the intellectual content of the manuscript, and approved the final version of the article to be published.
References
1. Korshunov A, Sycheva R, and Golanov A (2005) The prognostic relevance of molecular alterations in glioblastomas for patients age < 50 years Cancer 104(4) 825–832 https://doi.org/10.1002/cncr.21221 PMID: 15981281
2. Louis DN, Perry A, and Reifenberger G, et al (2016) The 2016 World Health Organization classification of tumors of the central nervous system: a summary Acta neuropathologica 131(6) 803–820 https://doi.org/10.1007/s00401-016-1545-1 PMID: 27157931
3. Stupp R, Hegi ME, and Mason WP, et al (2009) Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial lancet oncol 10(5) 459–466 https://doi.org/10.1016/S1470-2045(09)70025-7 PMID: 19269895
4. Wick W, Weller M, and van den Bent M, et al (2014) MGMT testing–the challenges for biomarker-based glioma treatment Nat Rev Neurol 10(7) 372–385 https://doi.org/10.1038/nrneurol.2014.100 PMID: 24912512
5. Felsberg J, Thon N, and Eigenbrod S, et al (2011) Promoter methylation and expression of MGMT and the DNA mismatch repair genes MLH1, MSH2, MSH6 and PMS2 in paired primary and recurrent glioblastomas Int J Cancer 129(3) 659–670 https://doi.org/10.1002/ijc.26083 PMID: 21425258
6. Hegi ME, Diserens AC, and Godard S, et al (2004) Clinical trial substantiates the predictive value of O-6-methylguanine-DNA methyltransferase promoter methylation in glioblastoma patients treated with temozolomide Clin Cancer Res 10(6) 1871–1874 https://doi.org/10.1158/1078-0432.CCR-03-0384 PMID: 15041700
7. Hegi ME, Diserens AC, and Gorlia T, et al (2005) MGMT gene silencing and benefit from temozolomide in glioblastoma N Engl J Med 352(10) 997–1003 https://doi.org/10.1056/NEJMoa043331 PMID: 15758010
8. Chinot OL, Wick W, and Mason W, et al (2014) Bevacizumab plus radiotherapy-temozolomide for newly diagnosed glioblastoma N Engl J Med 370(8) 709–722 https://doi.org/10.1056/NEJMoa1308345 PMID: 24552318
9. Gilbert MR, Dignam JJ, and Armstrong TS, et al (2014) A randomized trial of bevacizumab for newly diagnosed glioblastoma N Engl J Med 370(8) 699–708 https://doi.org/10.1056/NEJMoa1308573 PMID: 24552317 PMCID: 4201043
10. Wick W, Platten M, and Meisner C, et al (2012) Temozolomide chemotherapy alone versus radiotherapy alone for malignant astrocytoma in the elderly: the NOA-08 randomised, phase 3 trial Lancet Oncol 13(7) 707–715 https://doi.org/10.1016/S1470-2045(12)70164-X PMID: 22578793
11. Malmstrom A, Gronberg BH, and Marosi C, et al (2012) Temozolomide versus standard 6-week radiotherapy versus hypofractionated radiotherapy in patients older than 60 years with glioblastoma: the Nordic randomised, phase 3 trial Lancet Oncol 13(9) 916–926 https://doi.org/10.1016/S1470-2045(12)70265-6 PMID: 22877848
12. Rodriguez FJ, Thibodeau SN, and Jenkins RB, et al (2008) MGMT immunohistochemical expression and promoter methylation in human glioblastoma Appl Immunohistochem Mol Morphol 16(1) 59–65
13. Louis DN, Perry A, and Reifenberger G, et al (2015) The 2016 World Health Organization classification of tumors of the central nervous system: a summary Acta neuropathologica 131(6) 803–820 https://doi.org/10.1007/s00401-016-1545-1
14. Zhang FF, Morabia A, and Carroll J, et al (2011) Dietary patterns are associated with levels of global genomic DNA methylation in a cancer-free population J Nutr 141(6) 1165–1171 https://doi.org/10.3945/jn.110.134536 PMID: 21525250 PMCID: 3095144
15. Brait M, Ford JG, and Papaiahgari S, et al (2009) Association between lifestyle factors and CpG island methylation in a cancer-free population Cancer Epidemiol Biomarkers Prev 18(11) 2984–2991 https://doi.org/10.1158/1055-9965.EPI-08-1245 PMID: 19861513 PMCID: 2878974
16. Yang H, Wei D, and Yang K, et al (2014) The prognosis of MGMT promoter methylation in glioblastoma patients of different race: a meta-analysis Neurochem Res 39(12) 2277–2287 https://doi.org/10.1007/s11064-014-1435-7 PMID: 25230908
17. Ostrom QT, Gittleman H, and Farah P, et al (2013) CBTRUS statistical report: primary brain and central nervous system tumors diagnosed in the United States in 2006–2010 Neuro Oncol 15(Suppl 2) ii1–ii56 https://doi.org/10.1093/neuonc/not151 PMCID: 3798196
18. Chakrabarti I, Cockburn M, and Cozen W, et al (2005) A population-based description of glioblastoma multiforme in Los Angeles County, 1974–1999 Cancer 104(12) 2798–2806 https://doi.org/10.1002/cncr.21539 PMID: 16288487
19. Huse JT, Holland E, and DeAngelis LM (2013) Glioblastoma: molecular analysis and clinical implications Annu Rev Med 64 59–70 https://doi.org/10.1146/annurev-med-100711-143028
20. Krakstad C and Chekenya M (2010) Survival signalling and apoptosis resistance in glioblastomas: opportunities for targeted therapeutics Mol Cancer 9 135 https://doi.org/10.1186/1476-4598-9-135 PMID: 20515495 PMCID: 2893101
21. Hegi ME, Liu L, and Herman JG, et al (2008) Correlation of O6-methylguanine methyltransferase (MGMT) promoter methylation with clinical outcomes in glioblastoma and clinical strategies to modulate MGMT activity J Clin Oncol 26(25) 4189–4199 https://doi.org/10.1200/JCO.2007.11.5964 PMID: 18757334
22. Rivera AL, Pelloski CE, and Gilbert MR, et al (2010) MGMT promoter methylation is predictive of response to radiotherapy and prognostic in the absence of adjuvant alkylating chemotherapy for glioblastoma Neuro Oncol 12(2) 116–121 https://doi.org/10.1093/neuonc/nop020 PMID: 20150378 PMCID: 2940581
23. Esteller M, Hamilton SR, and Burger PC, et al (1999) Inactivation of the DNA repair gene O6-methylguanine-DNA methyltransferase by promoter hypermethylation is a common event in primary human neoplasia Cancer Res 59(4) 793–797 PMID: 10029064
24. Uno M, Oba-Shinjo SM, and Camargo AA, et al (2011) Correlation of MGMT promoter methylation status with gene and protein expression levels in glioblastoma Clinics (Sao Paulo) 66(10) 1747–1755 https://doi.org/10.1590/S1807-59322011001000013
25. Sanchez-Navarro I, Gamez-Pozo A, and Gonzalez-Baron M, et al (2010) Comparison of gene expression profiling by reverse transcription quantitative PCR between fresh frozen and formalin-fixed, paraffin-embedded breast cancer tissues Biotechniques 48(5) 389–397 https://doi.org/10.2144/000113388 PMID: 20569212
26. Ginzinger DG (2002) Gene quantification using real-time quantitative PCR: an emerging technology hits the mainstream Exp Hematol 30(6) 503–512 https://doi.org/10.1016/S0301-472X(02)00806-8 PMID: 12063017
27. Felsberg J, Malzkorn B, and Bujan B (2013) Molecular diagnostics of glioma—results of the first interlaboratory comparison of MGMT promoter methylation testing at twenty-three academic centers in Germany, Austria and the Netherlands Clin Neuropathol 32(414) e415
28. Van Krieken J, Siebers A, and Normanno N, et al (2013) European consensus conference for external quality assessment in molecular pathology Ann oncol 24(8) 1958–1963 https://doi.org/10.1093/annonc/mdt153 PMID: 23613479
29. Bady P, Sciuscio D, and Diserens AC, et al (2012) MGMT methylation analysis of glioblastoma on the Infinium methylation BeadChip identifies two distinct CpG regions associated with gene silencing and outcome, yielding a prediction model for comparisons across datasets, tumor grades, and CIMP-status Acta Neuropathol 124(4) 547–560 https://doi.org/10.1007/s00401-012-1016-2 PMID: 22810491 PMCID: 3444709
30. Kanemoto M, Shirahata M, and Nakauma A, et al (2014) Prognostic prediction of glioblastoma by quantitative assessment of the methylation status of the entire MGMT promoter region BMC cancer 14(1) 1 https://doi.org/10.1186/1471-2407-14-641






