Chromosomal rearrangements in acute myeloid leukemia (AML)
E Belloni1,2, M Trubia1,2, P Gasparini1,2, C Micucci1,2, C Tapinassi1,2, S Confalonieri1,2, P Nuciforo1,2, B Martino3, F Lo-Coco4, PP Di Fiore1,2,5 and PG Pelicci1,2,5
1Istituto
Europeo di Oncologia, Milan, Italy
2Campus IFOM-IEO, Milan, Italy
3Divisione di Ematologia, Azienda Ospedaliera
Bianchi-Malacrino-Morelli, Reggio Calabria, Italy
4Università di Roma Tor Vergata, Department of Biopatologia e
Diagnostica per Immagini, Rome, Italy
5Dipartimento di Medicina, Chirurgia ed Odontoiatria, Universita'
degli Studi di Milano, Milan, Italy
Correspondence to E Belloni. Email: elena.belloni@ifom-ieo-campus.it
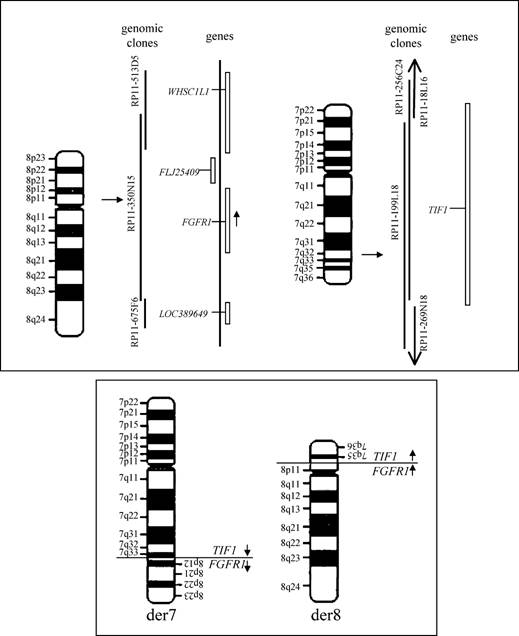
Chromosomal rearrangements in acute myeloid leukaemia (AML) frequently give rise to aberrant fusion proteins, responsible for the disease onset (50% of all reported cases). Five per cent of such cases harbour rare rearrangements, mostly translocations, resulting either in new rare fusion products or in the deregulation/truncation of specific genes. In this specific case, a new rearrangement between chromosomes 7 and 8 gives rise to a new abnormal fusion product between the FGFR1 and TIF1 genes.
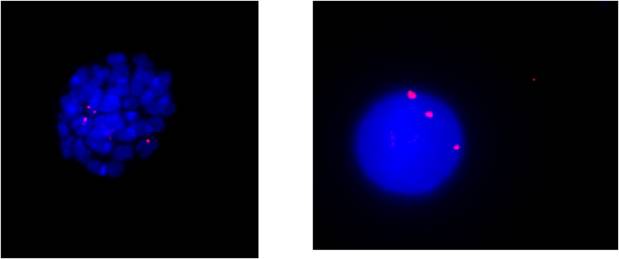
Fluorescence in situ hybridization (FISH) can be used to evidence the presence of such a rearrangement, by selecting genomic clones mapping in the chromosomal region that contains the breakpoint. The presence of the rearrangement can be detected in both interphase and metaphase nuclei. In the shown image, the clone 350N11, spanning the FGFR1 locus, has been labelled in red and used to hybridize interphase and metaphase nuclei (stained with DAPI, blue) on bone marrow cells derived from the patient. The presence of three signals indicates the clone is recognizing the normal chromosome 8, as well as the two derivative chromosomes 7 and 8, proving the presence of the breakpoint within this gene.





