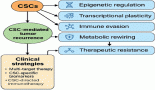
One of the most promising avenues for interpreting large datasets of molecular expression profiles involves pathway-based analysis. Pathways are collection of genes and proteins that perform a well-defined biological task. These pathways have been established through decades of molecular biology research and are collected in a variety of public pathway repositories (KEGG and Reactome Pathway database). Understanding the complexity of these pathways is critical for understanding normal biological conditions and disease states and also since the number of known pathways within the cells is significantly smaller than the number of genes that is typically profiled, the transformation of data from a gene-centric view to a pathway-centred one represents a dramatic reduction in the number of dimensions. Such reduction allows a biologist to interpret and understand the data in a manner that is not possible when it is viewed as a collection of individual genes.