Analysis of mutational variations in TP53 tumour suppressor gene among Pakistani head and neck cancer patients
Summera Fatima1, Asia Bibi1, Sara Samad Qureshi2 and Suman Khan2
1Department of Zoology, The Women University Multan, Multan 60000, Pakistan
2Nishtar Medical University & Hospital, Multan 60000, Pakistan
Abstract
The aim of this study was to determine the frequency of TP53 mutation among Pakistani head and neck cancer (HNC) patients who visited Nishtar Hospital Multan and Nishtar Institute of Dentistry (NID), Multan, Pakistan. While significant research has been conducted on the role of p53 in HNC throughout the world, this study is the first of its kind in Southern Punjab, Pakistan. A total of 242 samples (121 cases and 121 controls) were collected from Nishtar Hospital Multan and NID, Multan, Pakistan. After histopathological analysis to determine the stage type and grade of malignancy, DNA extraction and sequencing were carried out to assess any mutations in the TP53 region (exons 5–8). Genetic screening was performed by the polymerase chain reaction (PCR)-single strand conformation polymorphism (SSCP) technique and Chromas 2.6.6 was used to visualise the sequencing results. The mean age of cases was 50.73 ±16.41 years and controls were 37.55 ± 15.51 years. The frequency of HNC was higher in male patients (65.28%) than in female patients (34.71%). Overall, this cancer was found to be significantly more prevalent in the age group of >35–55 years (p < 0.001). Smoking (51% versus 14%), naswar usage (15.7% versus 6.6%), poor oral hygiene (52.9% versus 29.8%) and anemic status (57.0% versus 4.1%) were significantly associated with cases (p ≤ 0.05) compared to controls. Only 04 samples exon 5 (1 sample), exon 7 (2 samples) and exon 8 (1 sample) with changed mobility patterns were found on the SSCP gel. All mutations were missense and heterozygous. Out of four mutant samples, three mutations were in the hotspot regions (codon 175, 245 and 248) of p53. Based on this study, there may be a weak association between the TP53 exon 5–8 mutation and HNC patients in Southern Punjab, Pakistan.
Keywords: TP53, head and neck cancer, hotspot mutation, SSCP, missense mutation
Correspondence to: Asia Bibi
Email: drasia@wum.edu.pk
Published: 16/05/2024
Received: 14/01/2024
Publication costs for this article were supported by ecancer (UK Charity number 1176307).
Copyright: © the authors; licensee ecancermedicalscience. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Introduction
Head and neck cancer (HNC) accounts for 5% of all cancers globally and is the seventh most common cancer worldwide [1–5]; however, in Pakistan, HNC accounts for 32.6% of all cancers [65] and is considered as second most usual cancer [65, 66]. HNC prevalence is expected to increase by 30% yearly in 2030 in both advanced and poor countries [3, 4, 6]. Its prevalence is different because of anatomical sub sites and geographical differences. Smoking-linked HNC has been found to be decreased in developed countries and a rising trend has been found in Human papillomavirus (HPV) linked HNC but in developing countries, a continuous rise has been seen [67–70]. The highest ratio of HNC has been recorded in South East Asia and some countries of Central and South Europe such as France and Belgium. The frequency of different types of HNC is high in Pakistan, India, France, Brazil, Bangladesh, Afghanistan, Nepal, Sri Lanka, Iran, Maldives and Bhutan [71, 72].
Major causes of HNC include hereditary and natural harmful components such as tobacco, Epstein-Barr Virus, HPV, arecanut, liquor, eating less carbs and other ecological and occupational exposures [7, 8]. Other harmful factors are ecological exposures to wool dust, wood dust, mineral filaments and low consumption of vegetables and fruits [9, 10]. The most common modifications in HNSCC are the disturbance in the pathway of p53 [11, 12].
HNC is a complex illness that arises from changes in the tumour suppressor genes due to several gene-gene and gene-environment interactions. One of these genes is TP53, which is essential for controlling regular cell division and whose disturbance is the most frequent alteration in HNSCC (TP53; MIM#191170) [12]. In humans, the TP53 gene encodes the transcription factor tumour suppressor p53 protein. To maintain normal cellular functions and genome integrity, it is responsible for DNA repair, cellular homeostasis, metabolism, senescence and stimulation of non-apoptotic and apoptotic cell death. It is responsible for the activation of countless genes engaged in cell death metabolism and arrest of cell cycle in reaction to hypoxia, DNA damage, DNA mutations and oxidative stress, leading to stop the development of cancer [13]. So the loss of p53 activity leads to gene deletion or mutation, results in failure of normal cell activity to regulate and repair DNA death or damage, and finally causes cancer due to genomic variability [73].
TP53 mutations were confirmed in large-scale data in 2007 [14]. Almost 40%–60% of HNC patients have TP53 mutations leading to cancer progression from premalignant to invasive form and early recurrence of the disease is increased in these patients [15]. Point mutation makes the p53 protein non-functional or unstable, incapable of checking the proliferation of the cell. Non-functional p53 protein increases due to overexpression of the gene p53 and is reported in many studies [16]. Variations in p53 are common and appear early in HNC and both tobacco carcinogenic agents and HPV disease, two major risk factors, appear to target p53 [17]. The varying frequency (30% to 70%) of modifications in TP53 HNC patients in different studies is due to HNC heterogeneity (like cancer of various organs) and various methods used to detect TP53 mutational changes in HNC (such as mutational screening and immunohistochemistry). Many studies confirmed the role of TP53 mutations with a frequency range of 75% to 85% in non-HPV-associated HNSCC patients [18–21]. The majority of TP53 mutations shown by the International Agency for Research on Cancer affect exons 5–8, which make up the DNA-binding region specific to each site [22]. The goal of the current study was to determine if HNC patients from the Southern Punjab area had any mutations in the TP53 gene exon 5–8.
Materials and methods
In this study, we recruited a total of 242 participants, including 121 cases and 121 controls, from Nishtar Hospital Multan and Nishtar Institute of Dentistry, Multan, Pakistan, between 2017 and 2020. Informed consent was obtained from all the patients after informing them about the potential benefits of the study, in compliance with hospital rules. We collected Patient’s personal history, histopathological and other related information from the hospital and patients using a predesigned questionnaire. Ethical Approval for research work was obtained from the University Research Ethics Committee of Women University Multan (Reference No: WUM/UREC/00014).
The inclusion criteria for cases were HNC patients of any age (7 to 90 years), those not undergoing any treatment for cancer, and those with a clinical diagnosis of HNC. Fresh tissues and blood samples from 121 HNC cases were collected at the time of surgery before therapy. Various factors were compared between cases and controls including non-demographic factors (such as oral hygiene, diet, gum bleeding and anemic condition) and demographic factors (such as age, naswar use and smoking habit). The biopsy-proven HNC fresh tissues of any stage (TNM staging system includes tumour size and primary site, absence and presence of a metastatic condition of disease and nodal disease). The TNM stages are used to determine the overall stage of cancer, which ranges from 0 to IV [74], and were used in this investigation for the molecular analysis of the TP53 gene. To preserve the tissue, it was immediately put in liquid nitrogen and stored at −80°C. Following the manufacturer’s instructions, genomic DNA was isolated from fresh, frozen tissues of HNC using Thermo Scientific DNA Purification (Cat # K0721). Sequencing was carried out following Single Strand Conformation Polymorphism analysis (SSCP). SSCP was used to find mutational alterations in the core DNA binding domain of the TP53 gene, which contains important hot spots. Exon 5–8 regions of P53 were selected that give amplicons of sizes ranging from 180–247 base pairs. A mutant forward primer with a single nucleotide alteration by replacing T with G was chosen to provide the mutant positive control of exons 5 and 8. A DNA sequence altered from A to G was chosen as a mutation-positive control for exon 6, and a DNA sequence changed from G to C was chosen as a mutation-positive control for exon 7. Sequencing was used for the confirmation of all these mutant controls.
PCR cycling conditions were set at 95°C for 5 minutes, followed by 30 cycles of 30 seconds at 95°C, 45 seconds at the annealing temperature of the primers used (Exon 5 = 57°C, Exon 6 = 54°C, Exon 7 = 59°C and Exon 8 = 56°C), and 30 seconds at 72°C using the thermal cycler G-Storm 4822. The gel electrophoresis was conducted in 1× TAE buffer for 30 to 45 minutes at 100V followed by visualisation and photography using an ultraviolet transilluminator.
When Exon 5–8 of the TP53 gene were amplified then SSCP analysis was carried out on the Decode™ Universal Mutation Detection System, Biorad (Cat. # 170-9092). For the optimisation of SSCP of all four exons and gels were run in varying conditions, i.e., a. Exon 5 and 6: 60 mL of 12% non-denaturing polyacrylamide gel solution was prepared by mixing 24 mL of 30% acrylamide mix (29:1 acrylamide/bisacrylamide), 6 mL of 10× Tris boric acid EDTA (TBE) (pH 8.3) and 30 mL of deionized water and stored at 4°C. b. Exon 7 and 8: 60 mL of 12% non-denaturing polyacylamide gel solution was prepared by mixing 24 mL of 30% acrylamide mix (29:1 acrylamide/bisacrylamide), 33 mL of 10× TBE (pH 8.3) and 30 mL of deionized water and stored at 4°C.
For SSCP analysis, 5 µL PCR product was mixed with 15 µL of loading buffer (0.5 M NaOH, 0.05% bromophenol blue, 0.05% xylene cyanol and 50% (w/v) Sucrose) denatured at 96°C for 8 minutes and immediately placed on ice. Twenty microliters of each denatured PCR product (containing 30–40 ng DNA) were loaded onto a 12% gel. Electrophoresis was conducted with TBE running buffer (1× for exon 5 and 6: 0.5× for exon 7 and 8) at 220V for 10–12 hours until xylene cyanol reached the bottom of the gel. The temperature for electrophoresis was 20°C for exon 5 but for exon 6, 7 and 8, the best results were found at 4°C. After electrophoresis, the gels were separated from the plates using a gel separator and transferred to a tank containing an ethidium bromide solution (250 mL of deionized water and 250 uL of 10 mg/mL ethidium bromide solution) for 5 to 10 minutes. The gel was visualised on a UV transilluminator and photographed.
For results interpretation, we compared the SSCP band patterns of normal control, mutant control DNA and cases. No mutations were observed in control samples. Confirmation of all mutation-positive cases was done at least three times by a separate SSCP run and PCR reaction. Control samples did not show any mutation. Only those samples that exhibited a changed pattern in the form of extra bands and mobility shifts with respect to normal samples have proceeded for sequencing. These PCR products were amplified under the appropriate conditions, run on a 1.5% agarose gel and the bands exhibiting a changed pattern were cut to isolate DNA for sequencing using a Thermo Scientific GeneJET Gel Extraction kit (Cat. # K0691). Sequencing analysis was performed using the Sanger sequencing method and chromatograms were used to record sequencing results. The chromatograms were used to visually analyse the data and identify changes in the human TP53 gene in HNC patients.
The SPSS software, (version 20) was used to conduct statistical analyses. The demographic and non-demographic characteristics of cases and controls were assessed by the χ2 test. Multiple logistic regression was applied for multivariate analyses. We considered p-values of ≤0.05 as statistically significant.
Results
Out of the total 242 study participants, there was no difference in gender between the cases and controls, with an equal number of males (n = 79) and females (n = 42) in both groups. The mean age of the study population, ranged from 7 to 90 years, with cases having a mean age of 50.73 ± 16.41 years and controls having a mean age of 37.55 ± 15.51 years. A statistically significant difference (p ≤ 0.05) was observed among the groups with reference to demographic factors. HNC patients were significantly more frequent in the age group of >35–55 years (p < 0.001). HNC patients showed a high frequency of smoking (51% versus14%; p < 0.001), smoking frequency >10 times/day (90% versus 55%; p < 0.001), smoking duration >15 years (74% versus 5%; p < 0.001), naswar usage (15% versus 6%; p = 0.029), naswar use frequency 6–10 times/day (68% versus 50%; p = 0.370) and naswar use duration >20 years (52% versus 12%; p = 0.069) as compared to controls (Table 1).
Poor oral hygiene condition was significantly associated with cases (52% versus 29%; p = 0.026) as compared to controls but there was no significant association (57% versus 5%; p = 0.055), regarding gum bleeding factor between cases and controls. Anemic status was highly significantly associated with cases (57% versus 4%; p < 0.001) as compared to controls. With respect to diet, meat fonder was more frequent in cases as compared to controls (12% versus 3%; p = 0.055) (Table 2).
Only f4 out of 121 HNC samples (Exon 5, 7 and Exon 8) showed changed mobility patterns on the SSCP gel (Figure 1). All four mutant samples were from males. The affected site in Exon 5 and 7 was the larynx in three males, while in one male, the affected site (Exon 8) was the oral cavity. The patients had different occupations, with three being farmers and one being a cook. Tumour grade 2 was common in all four patients and all patients with the mutation had tumour stage III, with the affected site being the larynx, except for one patient who had tumour stage II, with the affected site being the oral cavity. Three of the patients smoked at the frequency of 20 times per day. Among the patients showing mutation, one was 45 years old and three patients were 60 years old (Table 3). Figure 2 shows the DNA sequencing chromatogram of mutations in Exon 5, 7 and 8. All exons sequencing chromatograms showed heterozygous mutations. In all four mutant samples, a missense mutation was observed. In Exon 5, a CGC > CAC (Arg > His) mutation was observed, and in Exon 7, a CGG > CAG (Gly > Ser) and CGG > CAG (Arg > Gln) mutations were observed and in Exon 8 a CGC > CAC (Arg > His) mutation was found. All four mutant samples had transition mutations. Mutations were found in the following codons; codon 175 (Exon 5), 245, 248 (Exon 7) and 283 (Exon 8) of the p53 gene (Table 4). The percentage of p53 mutation in Exon 5 and 8 was 0.82% and in Exon 7, it was 1.65%, while in Exon 6, it was 0% (Figure 3).
Table 1. Demographic features among cases and controls.
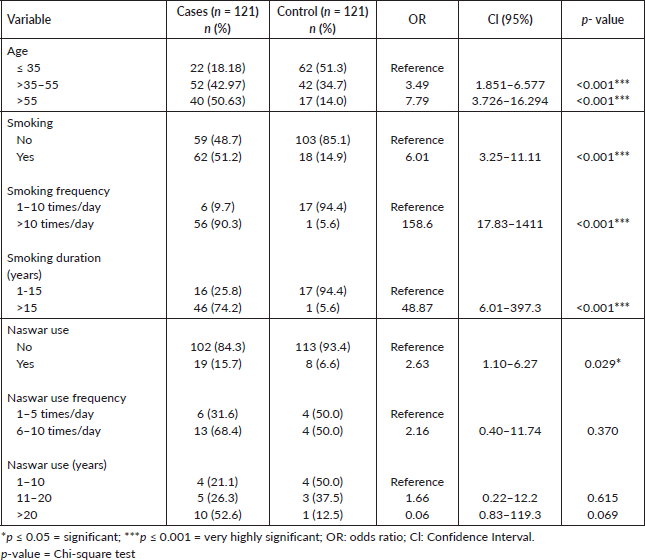
Table 2. Non-demographic factors among cases and controls.
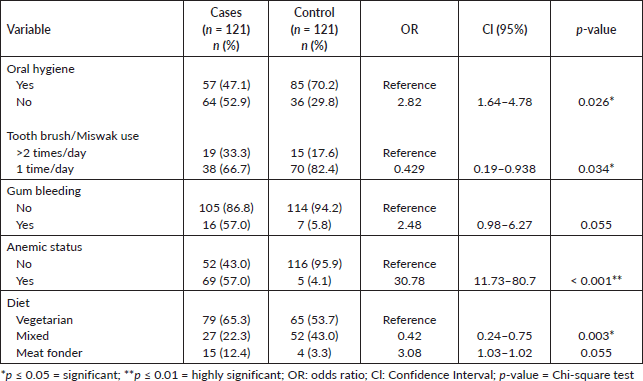
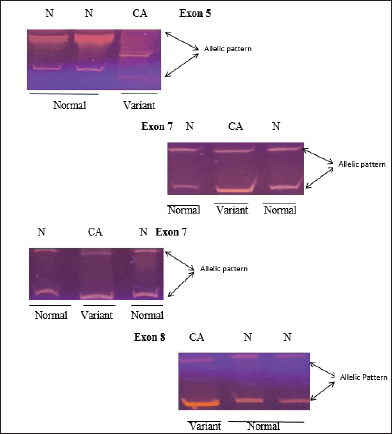
Figure 1. SSCP analysis of normal and variant mobility pattern. CA represents HNC patient while N indicates normal control gene amplified for exons 5, 7, and 8 of p53 gene.
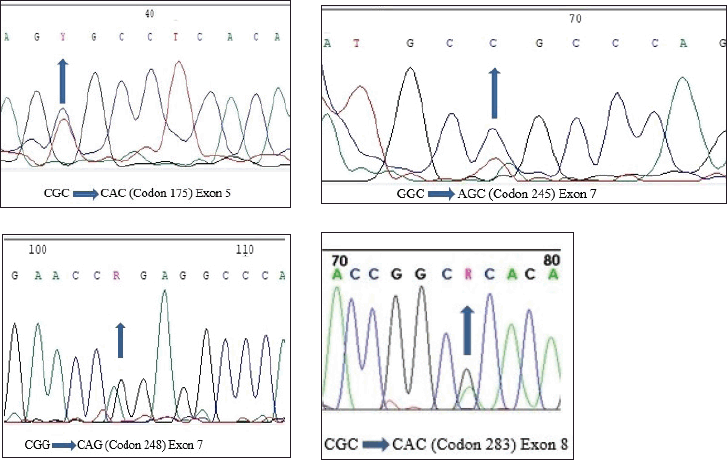
Figure 2. DNA sequencing chromatogram representing heterozygous mutations in each of exons 5, 7 and 8.
Table 3. Summary of mutant samples.

Table 4. Spectrum of TP53 mutations in HNC patients.
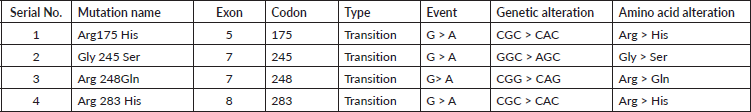
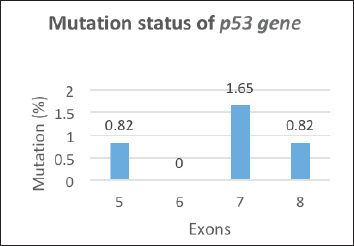
Figure 3. Mutation status of p53 gene.
Discussion
In this study, we compared demographic factors and non-demographic factors between cases and controls. The mean age of cases was 50.73(±16.41) years and controls were 37.55(±15.51) years. Similar mean age ranges were observed in different studies [23, 24], while in contrast, a mean age of 63.8 ± 9.3 years was found in both cases and control in a 2019 study [25]. A significant difference in demographic features was observed between cases and controls, which is consistent with one Pakistani study [26], while a contrary result was found in another Pakistani study [24]. Regarding smoking, the number of smokers was higher in cases compared to controls, as found in different studies [24, 27], but contrary to other studies [26, 25] where the frequency of smokers was higher in controls. In the current study, smoking frequency (>10 times/day) was more in cases than in controls, as found in a study conducted in 2020 [27]. Additionally, we observed a significant association in the case of naswar usage between cases and controls. The frequency of naswar use was higher in cases than in controls in this study, similar to a Pakistani study [24]. Therefore, naswar may be one of the risk factors for HNC, which is in agreement with one Pakistani study [28].
Oral health is crucial for overall health and well-being. Poor oral health can lead to various oral diseases. In this study, poor oral health conditions were more prevalent in cases than controls, consistent with other studies [29, 24].
Due to low literacy rates and limited income, people in this area neglect their oral health. Gum bleeding was observed more frequently in cases in line with other studies [30, 31, 23]. Our study found a significant association between cases and controls regarding anemic status and diet. The percentage of anemic patients was higher in cases as reported in one study [32] and more vegetables were consumed by cases than controls consistent with other studies [33, 34]. However, unlike our study, some studies have reported an inverse relationship between vegetable consumption and HNC [35–37]. Additionally, our study found that cases consumed less meat, whereas studies conducted in Asia reported an inverse relationship between red meat consumption and HNC risk [38].
In this study, the frequency of TP53 mutations was low (3.3%) which is similar to the findings of many studies [39–43] where it was 3.2%, 3%, 2%, 3.4% and 0%, respectively. However other studies have reported a higher frequency of TP53 mutations [44–48], where it was 24%, 25%, 21.5%, 16% and 66.2%, respectively. The reasons for the variation in TP53 mutation frequency observed in different studies may be due to differences in detection techniques and the number of exons (2–11 exons) analysed. Therefore, it is more challenging to draw conclusions that can be applied across studies from a comprehensive evaluation of the literature [46, 49, 50]. Nonetheless, our study supports the notion that the incidence of TP53 gene mutations differs globally, even within the same country.
In the current study, three mutant samples had affected site larynx (Exon 5 and 7) similar to different studies [51, 8] and one had affected site oral cavity (Exon 8), consistent with the study of Peltonen et al [51]. 75% of mutations were found in hotspot codons of p53, i.e., 175, 245 and 248 codons, as found in different studies [45, 51, 8, 52, 53]. No mutation was found in Exon 6 contrary to the finding of Peltonen
et al [51]. All mutations in the p53 gene found were missense mutations similar to different studies [51, 54, 8, 52, 55] that reported 90% of mutations in the p53 gene as missense mutations.
In the current study, all exon sequence chromatograms showed heterozygous mutations similar to the study of Leng et al [56] and contrary to the study of Baugh et al [52] which reported homosygous mutations in 50% to 60% of human cancer. Mutation in codon 245 was found in Exon 7, which matches with different studies [45, 58, 57, 8] that reported it as the most prevalent hotspot codon in HNC. The percentage of codon 245 mutation was 0.82%, compared to 0.4% in Baugh et al [52]. Mutation in codon 248 was found in Exon 7, similar to the study of Sisk et al [45], with a percentage of 0.82% and contrary to the study of Baugh et al [52], where codon 248% was 3.53%. In Exon 8, the mutation was found in codon 283 consistent with the study of Peltonen et al [51]. Mutation in codon 175 was found in Exon 5, similar to different studies [51, 52]. All four mutant samples showed only transition mutations contrary to the study of Schneider-Stock et al [58] where both transition and transversion mutations were observed. The age of patients was over 45 years old, consistent with the study of Poeta et al [14]. In this study with knowledge of the patients’ smoking habits and environmental exposure, we examined p53 aberrations in primary HNC patients. Three out of the four mutation-containing patients did smoking and were exposed to dust, insecticides and pesticides, similar to the factors observed in the study of Peltonen et al [51].
There is a well-established and significant link between smoking and HNC as found in several studies [59–63, 24, 25, 64] and it is known that p53 aberrations in general play a significant role in human cancers. Though the importance of p53 in the etiology of HNC has not yet been fully described in the literature, TP53 mutations in human malignancies have typically been the subject of substantial research. Mutant p53, the aberrant protein produced by TP53 alleles with missense mutations that frequently accumulate in cancer cells, has drawn a great deal of attention [51].
Conclusion
In the current study, we identified four mutations in exons 5–8 of the TP53 gene in HNC patients using SSCP. Our findings suggest that TP53 mutations may have a limited role in the development of HNC in the study population. However, further studies on a large scale are necessary to obtain more detailed results.
Acknowledgments
The authors acknowledge the participation of all HNC patients.
Conflicts of interest
The authors declare no conflict of interest.
Funding
No source of funding.
Author contributions
Summera Fatima: Data collection, data analysis, result compilation, writing – review, statistical analysis and editing and quality control.
Asia Bibi: Conceptualisation, methodology, writing – original draft, supervision.
Sara Samad Qureshi: Review, editing and data collection.
Suman Khan: Review, editing and data collection.
References
1. Cogliano VJ, Baan R, and Straif K, et al (2011) Preventable exposures associated with human cancers J Nat Cancer Inst 103(24) 1827–1839 https://doi.org/10.1093/jnci/djr483 PMID: 22158127 PMCID: 3243677
2. Dhull AK, Atri R, and Dhankhar R, et al (2018) Major risk factors in head and neck cancer: a retrospective analysis of 12-year experiences World J Oncol 9(3) 80–84 https://doi.org/10.14740/wjon1104w PMID: 29988794 PMCID: 6031231
3. Johnson DE, Burtness B, and Leemans CR, et al (2020) Head and neck squamous cell carcinoma Nat Rev Dis Prim 6(1) 92 https://doi.org/10.1038/s41572-020-00224-3 PMID: 33243986 PMCID: 7944998
4. Sung H, Ferlay J, and Siegel RL, et al (2020) Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries CA: Cancer J Clin 71(3) 209–249
5. Mody MD, Rocco JW, and Yom SS, et al (2021) Head and neck cancer Lancet 398(10318) 2289–2299 https://doi.org/10.1016/S0140-6736(21)01550-6 PMID: 34562395
6. Bravi F, Lee YA, and Hashibe M, et al (2021) Lessons learned from the INHANCE consortium: an overview of recent results on head and neck cancer Oral Dis 27(1) 73–93 https://doi.org/10.1111/odi.13502
7. Tumino R and Vicario G (2004) Head and neck cancers: oral cavity, pharynx, and larynx Epidemiol Prev 28(2 Suppl) 28–33 PMID: 15281603
8. Zhou G, Liu Z, and Myers JN (2016) TP53 mutations in head and neck squamous cell carcinoma and their impact on disease progression and treatment response J Cellular Biochem 9999 1–11
9. Mork J, Lie AK, and Glattre E, et al (2001) Human papillomavirus infection as a risk factor for squamous-cell carcinoma of the head and neck N Engl J Med 344 1125–1131 https://doi.org/10.1056/NEJM200104123441503 PMID: 11297703
10. Boccia S, Cadoni G, and Sayed-Tabatabaei FA, et al (2008) CYP1A1, CYP2E1, GSTM1, GSTT1, EPHX1 exons 3 and 4, and NAT2 polymorphisms, smoking, consumption of alcohol and fruit and vegetables and risk of head and neck cancer J Cancer Res Clin Oncol 134(1) 93–100 https://doi.org/10.1007/s00432-007-0254-5
11. Boutelle AM and Attardi LD (2021) p53 and tumor suppression: it takes a network Tren Cell Biol 31(4) 298–310 https://doi.org/10.1016/j.tcb.2020.12.011
12. Levine AJ (2022) Targeting the P53 protein for cancer therapies: the translational impact of P53 research Cancer Res 82(3) 362–364 https://doi.org/10.1158/0008-5472.CAN-21-2709 PMID: 35110395 PMCID: 8852246
13. Vousden KH and Prives C (2005) P53 and prognosis: new insights and further complexity Cell 120(1) 7–10 PMID: 15652475
14. Poeta ML, Manola J, and Goldwasser MA, et al (2007) TP53 mutations and survival in squamous-cell carcinoma of the head and neck N Engl J Med 357(25) 2552–2561 https://doi.org/10.1056/NEJMoa073770 PMID: 18094376 PMCID: 2263014
15. Brennan JA and Sidransky D (1996) Molecular staging of head and neck squamous carcinoma Cancer Metast Rev 15(1) 3–10 https://doi.org/10.1007/BF00049484
16. Homann N, Nees M, and Conradt C, et al (2001) Overexpression of p53 in tumor-distant epithelia of head and neck cancer patients is associated with an increased incidence of second primary carcinoma Clin Cancer Res 7(2) 290–296 PMID: 11234882
17. Toyooka S, Tsuda T, and Gazdar AF (2003) The TP53 gene, tobacco exposure and lung cancer Hum Mutat 21 229–239 https://doi.org/10.1002/humu.10177 PMID: 12619108
18. Agrawal N, Frederick MJ, and Pickering CR, et al (2011) Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1 Science 333 1154–1157 https://doi.org/10.1126/science.1206923 PMID: 21798897 PMCID: 3162986
19. Stransky N, Egloff AM, and Tward AD, et al (2011) The mutational landscape of head and neck squamous cell carcinoma Science 333 1157–1160 https://doi.org/10.1126/science.1208130 PMID: 21798893 PMCID: 3415217
20. Pickering CR, Zhang J, and Yoo SY, et al (2013) Integrative genomic characterization of oral squamous cell carcinoma identifies frequent somatic drivers Cancer Discov 3 770–781 https://doi.org/10.1158/2159-8290.CD-12-0537 PMID: 23619168 PMCID: 3858325
21. Cancer Genome Atlas Network (2015) Comprehensive genomic characterization of head and neck squamous cell carcinomas Nature 517 576–582 https://doi.org/10.1038/nature14129 PMID: 25631445 PMCID: 4311405
22. Hainaut P and Hollstein M (2000) P53 and human cancer: the first ten thousand mutations Adv Cancer Res 77 81–137 https://doi.org/10.1016/S0065-230X(08)60785-X
23. Chang JS, Lo HI, and Wong TY, et al (2013) Investigating the association between oral hygiene and head and neck cancer Oral Oncol 49(10) 1010–1017 https://doi.org/10.1016/j.oraloncology.2013.07.004 PMID: 23948049
24. Ahmed R, Malik S, and Khan MF, et al (2019) Epidemiological and clinical correlates of oral squamous cell carcinoma in patients from north-west Pakistan J Pak Med Assoc 69(8) 1074–1078 PMID: 31431755
25. Vučičević Boras V, Fučić A, and Baranović S, et al (2019) Environmental and behavioural head and neck cancer risk factors Cent Eur J Public Health 27(2) 106–109 https://doi.org/10.21101/cejph.a5565
26. Akhtar A, Hussain I, and Talha M, et al (2016) Prevalence and diagnostic of head and neck cancer in Pakistan Pak J Pharm Sci 29(5 Suppl) 1839–1846
27. Chang CP, Siwakoti B, and Sapkota A, et al (2020) Tobacco smoking, chewing habits, alcohol drinking and the risk of head and neck cancer in Nepal Inter J Cancer 147(3) 866–875 https://doi.org/10.1002/ijc.32823
28. Manzoor H, Ahmad N, and Shuja J, et al (2020) Epidemiology characteristic of head & neck cancers (HNCs) in Southwestern Pakistan: 21 years experience Children 402 1–70
29. Hashim D, Sartori S, and Brennan P, et al (2016) The role of oral hygiene in head and neck cancer: results from International head and neck cancer epidemiology (INHANCE) consortium Ann Oncol 27(8) 1619–1625 https://doi.org/10.1093/annonc/mdw224 PMID: 27234641 PMCID: 4959929
30. Moreno-Lopez LA, Esparza-Gomez GC, and Gonzalez-Navarro A, et al (2000) Risk of oral cancer associated with tobacco smoking, alcohol consumption and oral hygiene: a case-control study in Madrid, Spain Oral Oncol 36(2) 170–174 https://doi.org/10.1016/S1368-8375(99)00084-6 PMID: 10745168
31. Lissowska J, Pilarska A, and Pilarski P, et al (2003) Smoking, alcohol, diet, dentition and sexual practices in the epidemiology of oral cancer in Poland Eur J Cancer Prev 12(1) 25–33 https://doi.org/10.1097/00008469-200302000-00005 PMID: 12548107
32. Azria D, Zouhair A, and Serre A, et al (2005) Anémie et cancers des voies aérodigestives supérieures Bull Cancer 92(5) 445–451 PMID: 15932808
33. Zhou BF, Stamler J, and Dennis B, et al (2003) Nutrient intakes of middle-aged men and women in China, Japan, United Kingdom, and United States in the late 1990s: the INTERMAP study J Hum Hypertens 17(9) 623–630 https://doi.org/10.1038/sj.jhh.1001605 PMID: 13679952 PMCID: 6561109
34. AICR W (2007) Food, Nutrition, Physical Activity, and the Prevention of Cancer: A Global Perspective (Washington, DC: American Institute for Cancer Research)
35. Lagiou P, Talamini R, and Samoli E, et al (2009) Diet and upper-aerodigestive tract cancer in Europe: the ARCAGE study Inter J Cancer 124(11) 2671–2676 https://doi.org/10.1002/ijc.24246
36. Lucenteforte E, Garavello W, and Bosetti C, et al (2009) Dietary factors and oral and pharyngeal cancer risk Oral Oncol 45(6) 461–467 https://doi.org/10.1016/j.oraloncology.2008.09.002
37. Chuang SC, Jenab M, and Heck JE, et al (2012) Diet and the risk of head and neck cancer: a pooled analysis in the INHANCE consortium Cancer Causes Control: CCC 23(1) 69–88 https://doi.org/10.1007/s10552-011-9857-x
38. Tzoulaki I, Brown IJ, and Chan Q, et al (2008) Relation of iron and red meat intake to blood pressure: cross sectional epidemiological study BMJ (Clinical research ed.) 337 a258 https://doi.org/10.1136/bmj.a258 PMID: 18632704 PMCID: 2658466
39. Pinheiro NA and Villa LL (2001) Low frequency of p53 mutations in cervical carcinomas among Brazilian women Braz J Med Biol Res 34(6) 727–733 https://doi.org/10.1590/S0100-879X2001000600005 PMID: 11378660
40. Hedau S, Jain N, and Husain SA, et al (2004) Novel germline mutations in breast cancer susceptibility genes BRCA1, BRCA2 and p53 gene in breast cancer patients from India Breast Cancer Res Treat 88(2) 177–186 https://doi.org/10.1007/s10549-004-0593-8 PMID: 15564800
41. Aziz I (2011) Spectrum of Tp53 Tumor Suppressor Gene Mutations and Codon 72 Polymorphism In Pakistani Female Breast Cancer Patients [Doctoral Dissertation] (Lahore: University of the Punjab)
42. Yan B, Chen Q, and Xu J, et al (2020) Low-frequency TP53 hotspot mutation contributes to chemoresistance through clonal expansion in acute myeloid leukemia Leukemia 34(7) 1816–1827 https://doi.org/10.1038/s41375-020-0710-7 PMID: 31988438 PMCID: 7597970
43. Sadia H, Ullah M, and Irshad A, et al (2022) Mutational analysis of exons 5-9 of TP53 gene in breast cancer patients of Punjabi ethnicity Adv Life Sci 9(1) 18–23
44. Haraf DJ, Nodzenski E, and Brachman D, et al (1996) Human papilloma virus and p53 in head and neck cancer: clinical correlates and survival J Am Assoc Cancer Res 2(4) 755–762
45. Sisk EA, Soltys SG, and Zhu S, et al (2002) Human papillomavirus and p53 mutational status as prognostic factors in head and neck carcinoma Head Neck: J Sci Spec Head Neck 24(9) 841–849 https://doi.org/10.1002/hed.10146
46. Bosch FX, Ritter D, and Enders C, et al (2004) Head and neck tumor sites differ in prevalence and spectrum of p53 alterations but these have limited prognostic value Int J Cancer 111(4) 530–538 https://doi.org/10.1002/ijc.11698 PMID: 15239130
47. Golusinski P, Lamperska K, and Pazdrowski J, et al (2011) Analiza wystepowania mutacji w obrebie genu TP53 u chorych na raka płaskonabłonkowego głowy i szyi [Analysis of mutations within the TP53 gene in patients with squamous cell carcinoma of the head and neck] Otolaryngol Pol 65(2) 114–121 https://doi.org/10.1016/S0030-6657(11)70640-0 PMID: 21735667
48. Hashmi AA, Bukhari U, and Aslam M, et al (2023) Clinicopathological parameters and biomarker profile in a cohort of patients with head and neck squamous cell carcinoma (HNSCC) Cureus 15(7) e41941 PMID: 37588336 PMCID: 10425608
49. Komarova EA and Gudkov AV (2001) Chemoprotection from p53-dependent apoptosis: potential clinical applications of the p53 inhibitors Biochem Pharmacol 62(6) 657–667 https://doi.org/10.1016/S0006-2952(01)00733-X PMID: 11556286
50. Tandon S, Tudur-Smith C, and Riley RD, et al (2010) A systematic review of p53 as a prognostic factor of survival in squamous cell carcinoma of the four main anatomical subsites of the head and neck Cancer Epidemiol Biomark Prev 19(2) 574–587 https://doi.org/10.1158/1055-9965.EPI-09-0981
51. Peltonen JK, Helppi HM, and Pääkkö P, et al (2010) p53 in head and neck cancer: functional consequences and environmental implications of TP53 mutations Head Neck Oncol 2(1) 1–10 https://doi.org/10.1186/1758-3284-2-36
52. Baugh EH, Ke H, and Ke H, et al (2018) Why are there hotspot mutations in the TP53 gene in human cancers? Cell Death Differ 25(1) 154–160 https://doi.org/10.1038/cdd.2017.180 PMCID: 5729536
53. Srividya, Giridhar BH, and Vishwanath S, et al (2019) Profiling of germline mutations in major hotspot codons of TP53 using PCR-RFLP Pathol Oncol Res POR 25(4) 1373–1377 https://doi.org/10.1007/s12253-018-0394-8
54. Perri F, Pisconti S, and Scarpati GDV (2016) P53 mutations and cancer: a tight linkage Ann Transl Med 4 522 https://doi.org/10.21037/atm.2016.12.40
55. Huszno J and Grzybowska EWA (2018) TP53 mutations and SNPs as prognostic and predictive factors in patients with breast cancer (Review) Oncol Lett 16 34–40 PMID: 29928384 PMCID: 6006469
56. Leng K, Schlien S, and Bosch FX (2006) Refined characterization of head and neck squamous cell carcinomas expressing a seemingly wild-type p53 protein J Oral Pathol Med 35(1) 19–24 https://doi.org/10.1111/j.1600-0714.2005.00365.x PMID: 16393249
57. Thomas JOMD (2011) TP53 as a Biomarker in Head and Neck Squamous Cell Carcinoma The University of Texas MD Anderson Cancer Center UTHealth Graduate School of Biomedical Sciences [Dissertations and Theses (Open Access)] (Houston: The Texas Medical Center Library) p 217
58. Schneider-Stock R, Mawrin C, and Motsch C, et al (2004) Retention of the arginine allele in codon 72 of the p53 gene correlates with poor apoptosis in head and neck cancer Am J Pathol 164(4) 1233–1241 https://doi.org/10.1016/S0002-9440(10)63211-7 PMID: 15039212 PMCID: 1615339
59. Hashibe M, Brennan P, and Benhamou S, et al (2007) Alcohol drinking in never users of tobacco, cigarette smoking in never drinkers, and the risk of head and neck cancer: pooled analysis in the International head and neck cancer epidemiology consortium J Natl Cancer Inst 99(10) 777–789 https://doi.org/10.1093/jnci/djk179 PMID: 17505073
60. Hashibe M, Brennan P, and Chuang SC, et al (2009) Interaction between tobacco and alcohol use and the risk of head and neck cancer: pooled analysis in the International head and neck cancer epidemiology consortium Cancer Epidemiol Biomarkers Prev 18(2) 541–550 https://doi.org/10.1158/1055-9965.EPI-08-0347 PMID: 19190158 PMCID: 3051410
61. Koyanagi YN, Matsuo K, and Ito H, et al (2016) Cigarette smoking and the risk of head and neck cancer in the Japanese population: a systematic review and meta-analysis Jpn J Clin Oncol 46(6) 580–595 https://doi.org/10.1093/jjco/hyw027 PMID: 27369767
62. Lu Y, Sobue T, and Kitamura T, et al (2018) Cigarette smoking, alcohol drinking, and oral cavity and pharyngeal cancer in the Japanese: a population-based cohort study in Japan Eur J Cancer Prev 27(2) 171–179 https://doi.org/10.1097/CEJ.0000000000000283
63. Kanwal M, Haider G, and Zareef U, et al (2019) Addiction of tobacco chewing and smoking in the patients of head and neck squamous cell carcinoma: a descriptive epidemiological study in Pakistan Pak J Med Sci 35(6) 1712 https://doi.org/10.12669/pjms.35.6.1309 PMID: 31777521 PMCID: 6861502
64. Rupe C, Basco A, and Schiavelli A, et al (2022) Oral health status in patients with head and neck cancer before radiotherapy: baseline description of an observational prospective study Cancers 14(6) 1411 https://doi.org/10.3390/cancers14061411 PMID: 35326564 PMCID: 8945997
65. Hanif M, Zaidi P, and Kamal S, et al (2009) Institution-based cancer incidence in a local population in Pakistan: nine year data analysis Asian Pac J Cancer Prev 10 227–230 PMID: 19537889
66. Tariq A, Mehmood Y, and Jamshaid M, et al (2015) Head and neck cancers: incidence, epidemiological risk, and treatment options Int J Pharm Res Allied Sci 4(3) 21–34
67. Sturgis EM and Cinciripini PM (2007) Trends in head and neck cancer incidence in relation to smoking prevalence: an emerging epidemic of human papillomavirus-associated cancers Cancer 110(7) 1429–1435 https://doi.org/10.1002/cncr.22963 PMID: 17724670
68. Westra WH (2009) The changing face of head and neck cancer in the 21st century: the impact of HPV on the epidemiology and pathology of oral cancer Head Neck Pathol 3(1) 78–81 https://doi.org/10.1007/s12105-009-0100-y PMID: 20596995 PMCID: 2807531
69. Warnakulasuriya S (2009) Global epidemiology of oral and oropharyngeal cancer Oral Oncol 45(4-5) 309–316 https://doi.org/10.1016/j.oraloncology.2008.06.002
70. Vigneswaran N and Williams MD (2014) Epidemiologic trends in head and neck cancer and aids in diagnosis Oral Maxillofac Surg Clin North Am 26(2) 123–141 https://doi.org/10.1016/j.coms.2014.01.001 PMID: 24794262 PMCID: 4040236
71. de Camargo Cancela M, Voti L, and Guerra-Yi M, et al (2010) Oral cavity cancer in developed and in developing countries: population-based incidence Head Neck 32(3) 357–367 https://doi.org/10.1002/hed.21193
72. Khan Z, Tönnies J, and Müller S (2014) Smokeless tobacco and oral cancer in South Asia: a systematic review with meta-analysis J Cancer Epidemiol 2014 394696 https://doi.org/10.1155/2014/394696 PMID: 25097551 PMCID: 4109110
73. Muller PA and Vousden KH (2013) P53 mutations in cancer Nat Cell Biol 15 2–8 https://doi.org/10.1038/ncb2641
74. Deschler DG and Day T (2008) TNM staging of head and neck cancer and neck dissection classification Am Acad Otolaryn Head Neck Surg Found 10–23