Oxaliplatin: pre-clinical perspectives on the mechanisms of action, response and resistance
RN Seetharam, A Sood and S Goel
Department of Oncology, Montefiore Medical Center/Albert Einstein Cancer Center, 111 E 210th St, Bronx, NY 10467, USA
Correspondence RN Seetharam. Email: rneelavar@yahoo.com
Abstract
Oxaliplatin is a third-generation platinum compound that has shown a wide range of anti-tumour activity in metastatic cancer and in multiple cell lines. It contains a diaminocyclohexane carrier ligand and is one of the least toxic platinum agents. In the past decade, the use of oxaliplatin for the treatment of colorectal cancer has become increasingly popular because neither cisplatin nor carboplatin demonstrate significant activity. Similar to cisplatin, oxaliplatin binds to DNA, leading to GG intra-strand crosslinks. Oxaliplatin differs from its parent compounds in its mechanisms of action, cellular response and development of resistance, which are not fully understood. Like most chemotherapeutic agents, efficacy of oxaliplatin is limited by the development of cellular resistance. ERCC1 (excision repair cross-complementation group 1) mediated nucleotide excision repair pathway appears to be the major pathway involved in processing oxaliplatin, because the loss of mismatch repair does not lead to oxaliplatin resistance. Recent findings support the involvement of many genes and different pathways in developing oxaliplatin resistance. This mini-review focuses on the effects of oxaliplatin treatment on cell lines with special emphasis on colorectal cell lines.
Introduction
Colorectal cancer is the third leading cause of cancer-related mortality in men and women in the United States. It is estimated that 146,970 men and women will be diagnosed with, and 49,920 will die, of this cancer in 2009 [1]. The last three decades have witnessed a significant amount of basic research on platinum coordination complexes, leading to the pre-clinical screening of several thousand new molecules, of which only a few have entered clinical development. Although platinum drugs have a broad range of activity against malignant tumours, they are particularly active against germ cell tumours and epithelial ovarian cancer. In addition, they play a primary role in the treatment of small cell and non-small-cell lung, cervical, head and neck, colorectal and bladder cancer [2]. The platinum drugs such as cisplatin, carboplatin and oxaliplatin are used to treat a broad range of cancers; however, in most cases, their efficacy is limited by the development of resistance [3]. Due to this, the primary objective of researchers working in this area has been to identify compounds with superior efficacy, reduced toxicity, lack of cross-resistance or improved pharmacological characteristics as compared with the parent compound, cisplatin. Oxaliplatin (trans-L-1,2-diamino cyclohexane oxalatoplatinum) is a third generation platinum compound and the first platinum-based compound to show efficacy in the treatment of colorectal cancer [4] and approved for therapy as a front-line agent [5]. The intracellular targets and mechanisms of action/resistance of oxaliplatin differ from that of its predecessors, cisplatin and carboplatin. It is important to note that oxaliplatin is more active in colon cells [6], and that cisplatin-resistant cell lines are sensitive to oxaliplatin [7, 8].
Intracellular targets and mechanisms of action
Oxaliplatin and cisplatin are structurally distinct, but form the same types of adducts at the same sites on DNA [9–13]. In physiological conditions, oxaliplatin forms DNA adducts that are not at dynamic equilibrium [14]. Upon entering the cell, oxaliplatin first forms a transient monoadduct and then forms a stable diadduct, by mostly binding to the N(7) site of the guanine residues [15]. Intra-strand adducts are most abundant, and if not repaired, will block both DNA replication and transcription. Although platinum adducts can form inter-strand crosslinks by DNA–protein interaction, the proteinase resistant crosslinks are usually less than 1% of the total platinum adducts [16].
Oxaliplatin belongs to 1,2-diaminocyclohexane (DACH) carrier ligand family, whereas cisplatin and carboplatin belong to cis-diammine. There are some differences between compounds belonging to these families.
1. Bulkiness: DACH-Pt-DNA ligands are bulkier and more hydrophobic than cis-diammine-Pt-DNA and, perhaps, therefore, they are more effective in inhibiting DNA synthesis and are superior cytotoxic compounds [17].
2. Bond constraint: N–Pt–N bond angle is more constrained for DACH-Pt-DNA adducts than for cis-diammine-Pt-DNA adducts [18]. This might lead to slower mono-adduct to di-adduct conversion of DACH-Pt-DNA, leading to less stable adducts.
3. Computer modelling: the modelling indicated that this ring protrudes directly outward into and fills much of the narrowed major groove of the bound DNA, forming a markedly altered and less polar major groove in the area of the adduct. The differences in the structure of the adducts produced by cisplatin and oxaliplatin are consistent with the observation that they are differentially recognized by the DNA mismatch repair system, cisplatin being more easily recognized [11]. A detailed kinetic analysis of the insertion and extension steps of dNTP incorporation in the vicinity of the adduct shows that both DNA polymerase beta (pol beta) and DNA polymerase eta (pol eta) catalyse trans-lesion synthesis past oxaliplatin-GG adducts with greater efficiency than past cisplatin-GG adducts [19].
Oxaliplatin processing
Mismatch repair proteins, DNA damage-recognition proteins and trans-lesion DNA polymerases discriminate between Pt-GG adducts containing cis-diammine ligands (formed by cisplatin and carboplatin) and trans-RR-diaminocyclohexane ligands (formed by oxaliplatin) [19,20]. It is known that mismatch repair proteins, such as MutS and hMSH2 bind to cisplatin, but not to oxaliplatin adducts [21]. Loss of mismatch repair produces low levels of resistance to cisplatin but not oxaliplatin [22]. So, nucleotide excision repair pathway appears to be the major pathway involved in the processing of oxaliplatin.
Low levels of XPA, a protein involved in making a nick at the third end of the platinum adduct, in the tests tumour cell lines is sufficient to explain their poor ability to remove platinum adducts from DNA [23,24].
Nucleotide excision repair
ERCC1 and ERCC2 (xeroderma pigmentosa—XPD) are the two major genes involved in this pathway. It has been previously shown that the expression levels of the ERCC1 gene can significantly affect the ability of the drug to influence survival in patients with colon cancer [25].
The nucleotide excision repair (NER) reaction is carried out by a multi-enzyme complex and involves a stepwise process of recognition, incision, excision, repair synthesis and ligation [26,27]. ERCC1 along with XPF forms a critical heterodimer of the NER pathway because of its damage recognition creation of nick 5' to the lesion [26, 28–35]. XPF-ERCC1 is also known to be involved in recombinational DNA repair and in the repair of inter-strand crosslinks [36]. Figure 1 shows simplified steps in NER pathway.
Figure 1: DNA strand dual incision, 5' incision by ERCC1-XPF heterodimer is 22 nucleotide from lesion and 3' incision by XPG is six nucleotides from the lesion
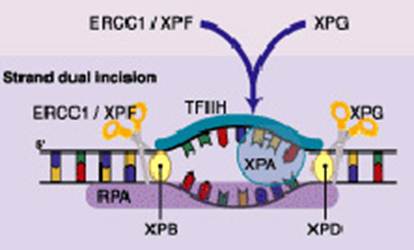
While there are indications that the relative ERCC1 mRNA level is a good marker for NER activity in human cancer cells, it is unclear whether expression of this gene has any relationship to other pathways of DNA repair [37]. In a study of 50 patients with ERCC1 gene expression ≤ 4.9 ´ 10(-3) (40 of 50 patients) had a median survival time of 10.2 months, compared with 1.9 months for patients with ERCC1 expression greater than 4.9 ´ 10(-3) (p < .001) [25].
Cellular response
There have been several studies on the cellular response of oxaliplatin in different types of cancer cell lines, sometimes suggesting contrasting results. In a study involving four cancer cell lines, ovarian and an inherently cisplatin-resistant colon (HT-29), ERCC1 mRNA levels measured after exposure to oxaliplatin for 20 hours were higher than in the control—the A2780 (ovarian) cell line [8]. Further, it was shown that, relative to cisplatin, a lower intracellular concentration and fewer DNA-Pt adducts are sufficient for oxaliplatin to exert its cytotoxicity [8]. Oxaliplatin is also capable of altering the voltage-gated sodium channels, thereby inducing both acute and chronic toxicity [38]. Another group studied the combination of irinotecan and oxaliplatin in HCT-8, a colorectal cancer cell line, and xenograft models and observed that ERCC1 expression was unregulated on exposure to oxaliplatin. Addition of irinotecan abrogated this effect, with the potential for synergy between the two drugs by the inhibition of DNA repair and increased cytotoxicity of the platinum [39]. In another study, it was shown that siRNA knockdown of ERCC1 expression resulted in sensitivity to oxaliplatin in the HeLa S3 (cervical cancer) cells [40].
Cytotoxicity of oxaliplatin on a panel of six colon cell lines in vitro showed that glutathione and glutathione S-transferase activity were not correlated to oxaliplatin cytotoxicity. Further, the expression of ERCC1 and XPA (xeroderma pigmentosum group A) demonstrated that ERCC1 expression was predictive of oxaliplatin sensitivity [41]. When DNA microarray analysis was used to analyse the transcriptional profile of resistant HCT116 colorectal cancer cells that were treated with oxaliplatin or 5-fluorouracil (5-FU), bioinformatic analyses identified sets of genes that were constitutively dysregulated in drug-resistant cells and transiently altered following acute exposure of parental cells to drug. This leads to the proposition that these genes may represent molecular signatures of sensitivity to oxaliplatin and 5-FU [42].
Resistance
The existing body of literature suggests that the rate of NER may have a major impact on the emergence of resistance and normal tissue tolerance to platinum drugs [43]. DNA adducts are differentially recognized by a number of cellular proteins. For example, mismatch repair proteins and some damage-recognition proteins bind to cisplatin-GG adducts with higher affinity than to oxaliplatin-GG adducts, and this differential recognition of cisplatin- and oxaliplatin-GG adducts is thought to contribute to the differences in cytotoxicity and tumour range of cisplatin and oxaliplatin [19]. Elevation of glutathione mediated by gamma-glutamyl transpeptidase has also been shown to be a mechanism of oxaliplatin resistance [8]. Oxaliplatin-resistance may also involve multiple other pathways like down-regulation of pyruvate kinase M2 [44], altered mitochondrial-mediated apoptosis [45] and phosphoinositide-3- kinase (PI3K)/Akt activation [46]. DNA microarray studies suggest the involvement of large number of genes in developing oxaliplatin resistance [42,47].
Conclusion and outlook
Although platinum drugs are one of the most widely used anti-cancer agents, the outcome of the treatment depends upon the drug resistance. Oxaliplatin has been shown to exhibit broad spectrum anti-tumour activity including a subset of cisplatin resistant cell lines. Pre-clinical studies have shown that ERCC1 gene expression plays critical role in the effectiveness of oxaliplatin treatment. Ongoing research will lead to the better understanding of the association between the expression levels of DNA excision repair genes and the response to oxaliplatin treatment. The goal of the ongoing research is to lead to the development of more effective compounds in this class.
References
1. American Cancer Society (2009) Cancer Facts & Figures—2009 Atlanta, Georgia, available from http://seer.cancer.gov/csr/1975_2006/results_single/sect_01_table.01.pdf
2. Lebwohl D and Canetta R (1998) Clinical development of platinum complexes in cancer therapy: an historical perspective and an update Eur J Cancer 34 1522–34 PMID: 9893623 doi: 10.1016/S0959-8049(98)00224-X
3. Martin LP, Hamilton TC and Schilder RJ (2008) Platinum resistance: the role of DNA repair pathways Clin Cancer Res 14 1291–5 PMID: 18316546 doi: 10.1158/1078-0432.CCR-07-2238
4. Meyerhardt JA and Mayer RJ (2005) Systemic therapy for colorectal cancer N Engl J Med 352 476–87 PMID: 15689586 doi: 10.1056/NEJMra040958
5. Goldberg RM, Sargent DJ, Morton RF, Fuchs CS, Ramanathan RK et al (2004) A randomized controlled trial of fluorouracil plus leucovorin, irinotecan, and oxaliplatin combinations in patients with previously untreated metastatic colorectal cancer J Clin Oncol 22 23–30 PMID: 14665611 doi: 10.1200/JCO.2004.09.046
6. Armand JP, Boige V, Raymond E, Fizazi K, Faivre S and Ducreux M (2000) Oxaliplatin in colorectal cancer: an overview Semin Oncol 27 96–104 PMID: 11049040
7. Rixe O, Ortuzar W, Alvarez M, Parker R, Reed E, Paull K and Fojo T (1996) Oxaliplatin, tetraplatin, cisplatin, and carboplatin: spectrum of activity in drug-resistant cell lines and in the cell lines of the National Cancer Institute's Anticancer Drug Screen panel Biochem Pharmacol 52 1855–65 PMID: 8951344 doi: 10.1016/S0006-2952(97)81490-6
8. Hector S, Bolanowska-Higdon W, Zdanowicz J, Hitt S and Pendyala L (2001) In vitro studies on the mechanisms of oxaliplatin resistance Cancer Chemother Pharmacol 48 398–406 PMID: 11761458 doi: 10.1007/s002800100363
9. Woynarowski JM, Chapman WG, Napier C, Herzig MC and Juniewicz P (1998) Sequence- and region-specificity of oxaliplatin adducts in naked and cellular DNA Mol Pharmacol 54 770–7 PMID: 9804612
10. Woynarowski JM, Faivre S, Herzig MC, Arnett B, Chapman WG et al (2000) Oxaliplatin-induced damage of cellular DNA Mol Pharmacol 58 920–7 PMID: 11040038
11. Scheeff ED, Briggs JM and Howell SB (1999) Molecular modeling of the intrastrand guanine-guanine DNA adducts produced by cisplatin and oxaliplatin Mol Pharmacol 56 633–43 PMID: 10462551
12. Wu Y, Pradhan P, Havener J, Boysen G, Swenberg JA, Campbell SL and Chaney SG (2004) NMR solution structure of an oxaliplatin 1,2-d(GG) intrastrand cross-link in a DNA dodecamer duplex J Mol Biol 341 1251–69 PMID: 15321720 doi: 10.1016/j.jmb.2004.06.066
13. Gelasco A and Lippard SJ (1998) NMR solution structure of a DNA dodecamer duplex containing a cis-diammineplatinum(II) d(GpG) intrastrand cross-link, the major adduct of the anticancer drug cisplatin Biochemistry 37 9230–9 PMID: 9649303 doi: 10.1021/bi973176v
14. Johnson NP, Hoeschele JD and Rahn RO (1980) Kinetic analysis of the in vitro binding of radioactive cis- and trans-dichlorodiammineplatinum(II) to DNA Chem Biol Interact 30 151–69 PMID: 7190076 doi: 10.1016/0009-2797(80)90122-2
15. Fichtinger-Schepman AM, van der Veer JL, den Hartog JH, Lohman PH and Reedijk J (1985) Adducts of the antitumour drug cis-diamminedichloroplatinum(II) with DNA: formation, identification and quantitation Biochemistry 24 707–13 PMID: 4039603 doi: 10.1021/bi00324a025
16. Zwelling LA, Anderson T and Kohn KW (1979) DNA-protein and DNA interstrand cross-linking by cis- and trans-platinum(II) diamminedichloride in L1210 mouse leukemia cells and relation to cytotoxicity Cancer Res 39 365–9 PMID: 570092
17. Raymond E, Chaney SG, Taamma A and Cvitkovic E (1998) Oxaliplatin: a review of preclinical and clinical studies Ann Oncol 9 1053–71 PMID: 9834817 doi: 10.1023/A:1008213732429
18. Gibbons GR, Page JD, Mauldin SK, Husain I and Chaney SG (1990) Role of carrier ligand in platinum resistance in L1210 cells Cancer Res 50 6497–501 PMID: 2208108
19. Chaney SG, Campbell SL, Bassett E and Wu Y (2005) Recognition and processing of cisplatin- and oxaliplatin-DNA adducts Crit Rev Oncol Hematol 53 3–11 PMID: 15607931 doi: 10.1016/j.critrevonc.2004.08.008
20. Sharma S, Gong P, Temple B, Bhattacharyya D, Dokholyan NV, and Chaney SG (2007) Molecular dynamic simulations of cisplatin- and oxaliplatin-d(GG) intrastand cross-links reveal differences in their conformational dynamics J Mol Biol 373 1123–40 PMID: 17900616 doi: 10.1016/j.jmb.2007.07.079
21. Farrell NP (2004) Preclinical perspectives on the use of platinum compounds in cancer chemotherapy Semin Oncol 31 1–9 PMID: 15726528 doi: 10.1053/j.seminoncol.2004.11.004
22. Manic S, Gatti L, Carenini N, Fumagalli G, Zunino F and Perego P (2003) Mechanisms controlling sensitivity to platinum complexes:role of p53 and DNA mismatch repair Curr Cancer Drug Targets 3 21–9 PMID: 12570658 doi: 10.2174/1568009033333727
23. Koberle B, Masters JR, Hartley JA and Wood RD (1999) Defective repair of cisplatin-induced DNA damage caused by reduced XPA protein in testicular germ cell tumours Curr Biol 9 273–6 PMID: 10074455 doi: 10.1016/S0960-9822(99)80118-3
24. Welsh C, Day R, McGurk C, Masters JR, Wood RD and Koberle B (2004) Reduced levels of XPA, ERCC1 and XPF DNA repair proteins in testis tumour cell lines Int J Cancer 110 352–61 PMID: 15095299 doi: 10.1002/ijc.20134
25. Shirota Y, Stoehlmacher J, Brabender J, Xiong YP, Uetake H et al (2001) ERCC1 and thymidylate synthase mRNA levels predict survival for colorectal cancer patients receiving combination oxaliplatin and fluorouracil chemotherapy J Clin Oncol 19 4298–304 PMID: 11731512
26. Sancar A (1996) DNA excision repair Annu Rev Biochem 65 43–81 PMID: 8811174 doi: 10.1146/annurev.bi.65.070196.000355
27. Petit C and Sancar A (1999) Nucleotide excision repair: from E. coli to man Biochimie 81 15–25 PMID: 10214906 doi: 10.1016/S0300-9084(99)80034-0
28. Altaha R, Liang X, Yu JJ and Reed E (2004) Excision repair cross complementing-group 1: gene expression and platinum resistance Int J Mol Med 14 959–70 PMID: 15547660
29. Park CH, Bessho T, Matsunaga T and Sancar A (1995) Purification and characterization of the XPF-ERCC1 complex of human DNA repair excision nuclease J Biol Chem 270 22657–60 PMID: 7559382 doi: 10.1074/jbc.270.39.22657
30. Biggerstaff M, Szymkowski DE and Wood RD (1993) Co-correction of the ERCC1, ERCC4 and xeroderma pigmentosum group F DNA repair defects in vitro EMBO J 12 3685–92 PMID: 8253090
31. Gillet LC and Scharer OD (2006) Molecular mechanisms of mammalian global genome nucleotide excision repair Chem Rev 106 253–76 PMID: 16464005 doi: 10.1021/cr040483f
32. Reardon JT and Sancar A (2005) Nucleotide excision repair Prog Nucleic Acid Res Mol Biol 79 183–235 PMID: 16096029 doi: 10.1016/S0079-6603(04)79004-2
33. Costa RM, Chigancas V, Galhardo Rda S, Carvalho H and Menck CF (2003) The eukaryotic nucleotide excision repair pathway Biochimie 85 1083–99 PMID: 14726015 doi: 10.1016/j.biochi.2003.10.017
34. Park CJ and Choi BS (2006) The protein shuffle. Sequential interactions among components of the human nucleotide excision repair pathway FEBS J 273 1600–8 PMID: 16623697 doi: 10.1111/j.1742-4658.2006.05189.x
35. van Hoffen A, Balajee AS, van Zeeland AA and Mullenders LH (2003) Nucleotide excision repair and its interplay with transcription Toxicology 193 79–90 PMID: 14599769 doi: 10.1016/j.tox.2003.06.001
36. Bergstralh DT and Sekelsky J (2008) Interstrand crosslink repair: can XPF-ERCC1 be let off the hook? Trends Genet 24 70–6 PMID: 18192062 doi: 10.1016/j.tig.2007.11.003
37. Reed E (1998) Platinum-DNA adduct, nucleotide excision repair and platinum based anti-cancer chemotherapy Cancer Treat Rev 24 331–44 PMID: 9861196 doi: 10.1016/S0305-7372(98)90056-1
38. Grolleau F, Gamelin L, Boisdron-Celle M, Lapied B, Pelhate M and Gamelin E (2001) A possible explanation for a neurotoxic effect of the anticancer agent oxaliplatin on neuronal voltage-gated sodium channels J Neurophysiol 85 2293–7 PMID: 11353042
39. Guichard S, Arnould S, Hennebelle I, Bugat R and Canal P (2001) Combination of oxaliplatin and irinotecan on human colon cancer cell lines: activity in vitro and in vivo Anticancer Drugs 12 741–51 PMID: 11593056 doi: 10.1097/00001813-200110000-00006
40. Chang IY, Kim MH, Kim HB, Lee DY, Kim SH, Kim HY and You HJ (2005) Small interfering RNA-induced suppression of ERCC1 enhances sensitivity of human cancer cells to cisplatin Biochem Biophys Res Commun 327 225–33 PMID: 15629453 doi: 10.1016/j.bbrc.2004.12.008
41. Arnould S, Hennebelle I, Canal P, Bugat R and Guichard S (2003) Cellular determinants of oxaliplatin sensitivity in colon cancer cell lines Eur J Cancer 39 112–9 PMID: 12504667 doi: 10.1016/S0959-8049(02)00411-2
42. Boyer J, Allen WL, McLean EG, Wilson PM, McCulla A, Moore S, Longley DB, Caldas C and Johnston PG (2006) Pharmacogenomic identification of novel determinants of response to chemotherapy in colon cancer Cancer Res 66 2765–77 PMID: 16510598 doi: 10.1158/0008-5472.CAN-05-2693
43. Gossage L and Madhusudan S (2007) Current status of excision repair cross complementing-group 1 (ERCC1) in cancer Cancer Treat Rev 33 565–77 PMID: 17707593 doi: 10.1016/j.ctrv.2007.07.001
44. Martinez-Balibrea E, Plasencia C, Gines A, Martinez-Cardus A, Musulen E et al (2009) A proteomic approach links decreased pyruvate kinase M2 expression to oxaliplatin resistance in patients with colorectal cancer and in human cell lines Mol Cancer Ther 8 771–8 PMID: 19372549 doi: 10.1158/1535-7163.MCT-08-0882
45. Gourdier I, Del Rio M, Crabbe L, Candeil L, Copois V et al (2002) Drug specific resistance to oxaliplatin is associated with apoptosis defect in a cellular model of colon carcinoma FEBS Lett 529 232–6 PMID: 12372606 doi: 10.1016/S0014-5793(02)03347-1
46. Leelawat K, Narong S, Udomchaiprasertkul W, Leelawat S and Tungpradubkul S (2009) Inhibition of PI3K increases oxaliplatin sensitivity in cholangiocarcinoma cells Cancer Cell Int 9 3 PMID: 19128511 doi: 10.1186/1475-2867-9-3
47. Samimi G, Manorek G, Castel R, Breaux JK, Cheng TC, Berry CC, Los G and Howell SB (2005) cDNA microarray-based identification of genes and pathways associated with oxaliplatin resistance Cancer Chemother Pharmacol 55 1–11 PMID: 15378272 doi: 10.1007/s00280-004-0819-9