Spectrum and management of breast cancer patients with variant of uncertain significance mutations at a tertiary care centre in North India
Abhenil Mittal1, S V S Deo2, Ajay Gogia1, Atul Batra1, Akash Kumar3, Sandeep Bhoriwal2, Koushik Sinha Deb4, Ekta Dhamija5, V L Ramprasad6, Olufunmilayo Olopade7 and Raja Pramanik1
1Department of Medical Oncology, All India Institute of Medical Sciences, New Delhi 110029, India
2Department of Surgical Oncology, All India Institute of Medical Sciences, New Delhi 110029, India
3Department of Medical Oncology, National Cancer Institute, Jhajjar, Haryana 110029, India
4Department of Psychiatry, All India Institute of Medical Sciences, New Delhi 110029, India
5Department of Radiodiagnosis, All India Institute of Medical Sciences, New Delhi 110029, India
6MedGenome Labs Ltd, Bengaluru, Karnataka 560099, India
7Center for Clinical Cancer Genetics and Global Health, University of Chicago, Chicago, IL 60637, USA
Abstract
Background: The spectrum and significance of Variants of Uncertain Significance (VUS) mutations in breast cancer predisposition genes is poorly defined in the Indian population.
Methods: All new female breast cancer patients from 1 March 2019 to 28 February 2020 were screened. Those providing informed consent and without previous genetic testing were recruited. Multigene panel testing (107 genes) by next-generation sequencing was performed for all patients. Descriptive statistics was used to describe the spectrum of VUS mutations.
Results: Out of 236 patients recruited in the study, a VUS was detected in 89 patients (37.71%). VUS pathogenic ratio was 2.02. A total of 121 different VUS mutations in 40 different genes were detected. Fourteen patients (15.7%) had a VUS in high penetrance genes and 36 VUS mutations (29.8%) were detected in one of the genes involved in homologous recombination repair pathway. No therapeutic interventions were done based on VUS.
Conclusions: In this large prospective study of genetic determinants of breast cancer from India, a high prevalence of VUS (37.71%) was detected with 15.7% patients having a VUS in high penetrance genes. More evidence needs to be generated from larger multicentric studies to better understand the implications of these genetic variants and enable their reclassification.
Keywords: variant of uncertain significance, next-generation sequencing, breast cancer, India
Correspondence to: Raja Pramanik
Email: drrajapramanik@gmail.com
Published: 01/08/2022
Received: 30/03/2022
Publication costs for this article were supported by ecancer (UK Charity number 1176307).
Copyright: © the authors; licensee ecancermedicalscience. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Introduction
Breast cancer is the most prevalent cancer worldwide and is also the most common cancer among Indian women. It also constitutes the major cause of mortality worldwide and in India [1, 2]. Approximately 10%–15% of breast cancers are hereditary and mutations in BRCA 1/2 account for half of the hereditary cancer burden (4%–6% of unselected breast cancer population) [3–7]. With the decreasing cost of genetic testing and increasing availability, the paradigm has shifted towards selecting patients who should not be offered testing, rather than selecting patients for germline testing [8]. Previous studies from large US based institutions showed that National Comprehensive Cancer Center Criteria (NCCN) were relatively insensitive to select patients for genetic testing and up to 50% patients with pathogenic/likely pathogenic mutations may be missed if only NCCN criteria are used to select patients. Work from our centre also showed that sensitivity of these criteria was suboptimal when applied to an Indian breast cancer population and we suggested relaxing the age cut off in India to include all women with breast cancer up to 60 years of age to achieve optimal sensitivity, however, this data does need validation in larger studies [9–12].
The optimum method of genetic testing still remains a matter of debate and depends on clinical setting, cost and availability [13]. However, in recent years, multi-gene panels have gained popularity and have become the standard of care for hereditary breast cancer at most institutions [14–16]. Advantages include saving both money and time, detection of many uncommon high penetrance genes and moderate penetrance genes for which evidence base is now accumulating and availability of comprehensive genomic information which can help in counselling patients and their families better and make informed therapeutic decisions [17, 18]. Multi-gene panels do come with their own set of issues including detection of pathogenic variants in genes for which management strategies are not well defined as yet and high rate of variants of uncertain significance (VUS).
VUS are defined by both the American College of Medical Genetics (ACMG) and International Agency for Research on Cancer as variants with scarce clinical information due to lack of population level evidence, scarce functional evidence, different evaluation and interpretation by clinicians and researchers and being usually found in non-coding regions as mis-sense or synonymous substitutions [13, 19, 20]. Unfortunately, VUS are the most frequently detected variants by next-generation sequencing (NGS), especially in high-risk genes like BRCA1/2, accounting for >40% of all variants detected with pathogenic/likely pathogenic variants accounting for <20% [19, 21]. The ever-growing accumulation of genetic data generates larger and larger percentages of VUS, and this is especially true of oncological diseases, for which large gene-panel sequencing is often required [22, 23]. It is expected that the frequency of VUS will decrease as more data accumulates from various races and ethnic backgrounds.
Majority of data on VUS in various international databases are derived from studies in western population [24, 25]. Data regarding VUS from India has been hardly reported in previous studies reflecting an urgent need for incorporating data from Indian patients in these databases [26, 27]. Therefore, in this study, we report on the spectrum of VUS mutations in consecutive breast cancer patients treated at an Indian tertiary care centre.
Methods
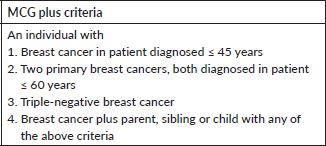
We screened all new breast cancer patients above the age of 18 years registered at our Institute’s breast cancer clinic over a 1-year period for participation in the study. Patients who had undergone previous germline genetic testing or who did not consent were excluded. After recording a detailed clinical history in a predesigned proforma, a three-generation pedigree chart was built based on family history. American Joint Committee on Cancer 8th edition was used to stage the patients. All patients were grouped into those qualifying the NCCN 2018 criteria or Mainstreaming Cancer Genetics (MCG) Plus criteria [28] for testing and those who do not using a predesigned checklist (Supplementary Tables 1 and 2). After the patients had been through pre-test genetic counselling by a medical oncologist, a 5 ml peripheral blood draw was made at the first visit and subjected to targeted NGS using the TruSight Cancer Sequencing Panel (Illumina, USA). The panel covered 107 high-risk genes known to be associated with cancer predisposition and all testing was performed at a centralised College of American Pathologists and Clinical Laboratory Improvement Amendments certified laboratory. Reflex multiplex ligation by probe amplification (MLPA method) was done to identify large genomic rearrangements in BRCA1 and 2 genes in cases where NGS did not reveal a pathogenic mutation.
Variants thus obtained by NGS/MLPA were grouped as benign, likely benign, pathogenic, likely pathogenic and VUS according to the ACMG Guidelines (Figure 1). Sanger sequencing was performed to confirm pathogenic/likely pathogenic variants. The study was approved by the Institute Ethics Committee (IEC) and was done according to good clinical practise guidelines as outlined in the Declaration of Helsinki.
Technical details of NGS and variant interpretation
From 50 ng of input genomic DNA of each patient, NGS libraries were prepared and hybridised to a custom pool of oligonucleotides, targeting genomic regions followed by paired end sequencing of up to 150 bp read lengths. DNA extracted from blood was used to perform targeted gene capture using a custom capture kit. The libraries were sequenced to mean > 80–100× coverage on Illumina sequencing platform. The sequences obtained were aligned to human reference genome (GRCh37/hg19) using the Burrows Wheeler Aligner (BWA) program and analysed using Picard and the Genome Analysis Toolkit (GATK) version 3.6 to identify variants relevant to the clinical indication. The GATK best practices framework was followed for identification of variants in the sample. Gene annotation of the variants was performed using the Variant Effect Predictor (VEP) program against the Ensembl release 87 human gene model. Clinically relevant mutations were annotated using published variants in literature and a set of diseases databases – ClinVar, OMIM, GWAS, HGMD and SwissVar. Common variants were filtered based on allele frequency in 1000 Genome Phase 3, ExAC, EVS, dbSNP147, 1000 Japanese Genome and our internal Indian population database. Non-synonymous variants effect was calculated using multiple algorithms such as PolyPhen-2, Scale Invariant Future Transform (SIFT), Mutation Taster2, Mutation Assessor and Likelihood Ratio Test (LRT). Only non-synonymous and splice site variants found in the hereditary cancer gene panel were used for clinical interpretation. Genetic test results were reported based on the recommendations of ACMG.
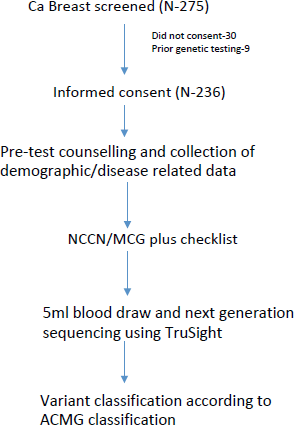
Figure 1. Study schema.
Results
Baseline characteristics between patients with and without VUS
A total of 236 breast cancer patients were recruited in the study out of 275 that were screened. Nine patients (3.3%) had previous germline testing and 30 patients (10.9%) did not consent.
Out of these 236 patients, a VUS was detected in 89 patients (37.71%). Mean age of this cohort was 46.2 years (18–69 years) and this was similar to patients who did not have a VUS (Table 1). Among patients with VUS, 5 patients (5.6%) had stage I disease, 27 patients (30.3%) had stage II disease and 31 (34.8%) and 26 patients (29.2%) had stage III and metastatic disease, respectively. The stage distribution was similar in patients with and without VUS. Patients with VUS were more often Her2 positive (42.7% versus 27.9%, p = 0.019) but significant family history was seen less often (8% versus 22.5%, p = 0.026). Rates of triple negative breast cancer (30.3% versus 38%) were similar between patients irrespective of VUS. Other baseline characteristics are detailed in Table 1.
VUS characteristics
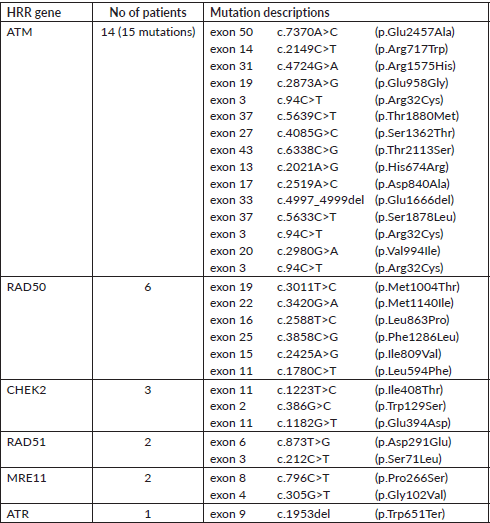
A total of 121 VUS mutations were seen in 40 different genes among this cohort. Sixty three patients had a single VUS mutation, two patients had two mutations in different genes, whereas six patients had three simultaneous mutations in three different cancer predisposition genes. All different genes with number of mutations in each gene are tabulated in Supplementary Table 3. Interestingly, there were 14 patients (15.7%) with 15 VUS mutations (12.4%) in high penetrance genes (BRCA1-3, BRCA2-4, CDH1-5 and TP53-3) (Table 2). However, among them, only one patient had a significant family history of breast cancer in first-degree relative; however, she is on routine surveillance. A total of 36 (29.8%) VUS mutations were seen in other homologous recombination repair (HRR) pathway genes (ATM-15, RAD50-6, BRCA2-4, BRCA1-3, CHEK2-3, RAD51-2, MRE11-2, ATR-1) (Table 3).
Table 1. Baseline characteristics.
Spectrum of pathogenic mutations seen in 44 patients has been previously reported giving a VUS/pathogenic ratio of 2.02 [12]. The VUS pathogenic ratio for high penetrance genes was 0.38 (13/34). None of the patients with VUS underwent any risk reducing interventions or family testing. No variants were reclassified as pathogenic/likely pathogenic (P/LP) till last follow-up.
Discussion
In this prospective study on unselected consecutive breast cancer patients at a north Indian tertiary care centre, all of whom underwent multigene hereditary germline panel testing, we found a high rate of VUS mutations (37.71%), although the VUS to pathogenic ratio was low (2.02) owing to a high rate of P/LP mutations in the cohort. More significantly, nearly one third of the mutations were seen in HRR pathway genes (29.8%) with 14 patients (15.7%, BRCA1/2 – 7.9%) having VUS mutations in high penetrance genes further complicating the decision-making process.
Since VUS is defined as a variant with a possibility of association with cancer occurrence between 5.1% and 94.9%, it is apparent that this wide definition accounts for the wide variation in reported prevalence and interpretation. In a large population based study on more than 2,000 women by Tung et al [29] across multiple centres in the US, a VUS rate of 41.6% was reported. They tested a relatively young cancer population (median age of 47 years) with 97% having a personal history of breast cancer. This patient disposition was very similar to our study except it was done in a predominantly white population, which likely accounted for a much lower incidence of P/LP mutation and a higher VUS/pathogenic ratio. Similar rates of VUS were reported by Slavin et al [30] and Beitsch et al [10] in their large datasets from the US using broad multigene panels reflecting consistency of the data. On the other hand, Chong et al [31] and Yadav et al [9] found lower rates of VUS when a limited panel was used. Our reported rate of 37.7% is much closer to that reported by studies using broad gene panels as our testing panel had 104 genes. It can be argued that using only BRCA1/2 sequencing or limited gene panels could limit the detection of VUS, however, it is unlikely to eliminate it and that would mean missing out on potentially vast amount of genetic information that can be used to inform future decision-making. Moreover, running from the problem is seldom the solution and the more data we collect, the lesser VUS we are likely to find in the future.
Table 2. VUS mutations in high penetrance genes.
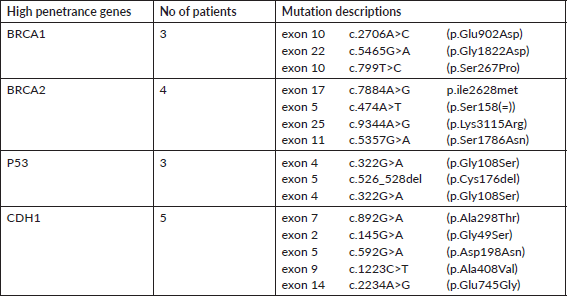
Table 3. VUS mutations in other HRR pathway genes (except BRCA1/2).
There is only one study from India by Singh et al [26] that has mentioned VUS rates in their cohort when testing by multigene panel. This was a laboratory-based study on women who were referred for germline testing. Although a 94-gene panel was used, data was reported for 14 high-risk genes and a VUS rate of 15% was found with a 30.1% P/LP mutation rate. A significant referral bias complicates interpretation of this data as VUS rates are generally higher than P/LP rates when using multigene panel testing as seen in large western studies. Our study provides the first prospectively and systematically collected information on VUS from India using a multigene panel and represents the first step in establishing a genetic registry of VUS. We found a VUS/pathogenic ratio of 2.02 which is similar to reported by other large western studies as we tested consecutive unselected patients [9, 10, 29, 30].
Although VUS in any gene complicates decision-making for clinicians and is a source of anxiety for patients, VUS in high penetrance cancer causing genes like BRCA1/2 is especially problematic [32, 33]. Variable VUS rates in BRCA1/2 genes have been reported in various studies ranging from 3.6% in study from US, 6.25% in a study from India, 10.6% from Nigeria and 23.1% from UK (only 52 patients studied) [26, 33–35]. In our cohort, seven patients had a BRCA1/2 VUS (7.9%), similar to that previously seen by Chheda et al [27]. Only one patient [36] had a significant family history of breast cancer in her mother and maternal grandmother. Currently, guidelines do not recommend therapeutic decisions based on VUS and these patients should be followed up with routine surveillance [19, 37, 38]. Highlighting the dynamics of complex decision-making in patients with VUS, Welsh et al [39] published their experience with 97 patients, all of whom had a BRCA VUS. Interestingly, 22% of these patients underwent a contralateral prophylactic mastectomy (CPM). However, these rates were not different from rates of CPM in other breast cancer patients without a VUS (25%) reflecting patient choice rather than effect of VUS [39]. Similar findings were reported by Chang et al [37] who found a VUS in BRCA1/2 in 5.7% of patients but having a VUS did not predict for prophylactic interventions. In our study as well, none of the patients with VUS had any therapeutic interventions in concordance with international guidelines.
However, in the absence of Indian data on genetic mutations, the reliance is on western mutation data which can be problematic. The spectrum and prevalence of P/LP mutations reported from the Indian population is quite different from the west and it stands to reason that same will be the case with VUS [12, 26]. Therefore, establishing a registry of VUS mutations is an urgent need in Indian cancer genetics in order to enable reclassifications based on data generated from a racially and ethnically concordant cohort. This study is a first step in that direction.
In the above-mentioned study by Welsh et al [39], 95% of the BRCA VUS were eventually classified as benign; however, 5% were reclassified as P/LP. Slavin et al [40] reported that variant reclassification rates can go as high as 17% and can occur as late as 20 years from initial testing, reflecting need for detailed post-test counselling addressing these issues, continued reassessment and regular follow-up in the genetics clinic. As part of mainstreaming genetic testing at our centre, these patients have been under regular follow-up with their primary oncologists with plan to return to genetics clinic if there is reclassification of VUS to P/LP. From our cohort, no VUS has been upgraded till date.
Our study has some strengths and several limitations. This is the first study to provide comprehensive data on spectrum of VUS mutations from unselected Indian breast cancer patients and is likely to add to the growing body of literature on genetic variant information from India. However, a limited number of patients were eventually recruited due to exhaustion of funds and were from a single tertiary care centre which are definite limitations of the study.
Conclusion
To conclude, VUS remains a significant clinical problem in clinical cancer genetics and this has escalated with expanding access to genetic testing especially with multigene panels. We report a high percentage of VUS (37.7%) in an unselected breast cancer population from India with a VUS pathogenic ratio similar to western studies. This represents the first prospective systematically collected database of VUS mutations in Indian population. This study lays the foundation for further multicentric studies which are currently being planned which will contribute to building up of Indian genetic data registry. Going forward, this will be crucial for reducing the incidence of VUS and therapeutic decision-making.
Conflicts of interest
All the authors declare no potential conflicts of interest.
Ethical approval
The study protocol was approved by the IEC vide letter number: IECPG-35/23.01.2019.
Funding
TATA Trusts at the University of Chicago, New Delhi Center.
Consent to participate
Informed consent was obtained from all patients.
Availability of data and material
Data regarding this study will be available from the corresponding author (RP) on reasonable request.
Authors’ contributions
Authors (RP, OO, AM, SVSD, AG, AB, AK, SB, KSD, ST, and ED) contributed to the concept design and conduct of the study. RP and AM did genetic counselling, AM and RP did the statistical analysis. AM and RP drafted the manuscript and all authors edited the manuscript.
References
1. Bray F, Ferlay J, and Soerjomataram I, et al (2018) Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries CA Cancer J Clin 68(6) 394–424 https://doi.org/10.3322/caac.21492 PMID: 30207593
2. Mathur P, Sathishkumar K, and Chaturvedi M, et al (2020) Cancer statistics, 2020: report from National Cancer Registry Programme, India JCO Glob Oncol [Internet] 6 1063–1075 [https://ascopubs.org/doi/full/10.1200/GO.20.00122] Date accessed: 4/12/20 https://doi.org/10.1200/GO.20.00122 PMID: 32673076 PMCID: 7392737
3. Claus EB, Risch N, and Thompson WD (1994) Autosomal dominant inheritance of early-onset breast cancer. Implications for risk prediction Cancer 73(3) 643–651 https://doi.org/10.1002/1097-0142(19940201)73:3<643::AID-CNCR2820730323>3.0.CO;2-5 PMID: 8299086
4. Peto J, Collins N, and Barfoot R, et al (1999) Prevalence of BRCA1 and BRCA2 gene mutations in patients with early-onset breast cancer J Natl Cancer Inst 91(11) 943–949 https://doi.org/10.1093/jnci/91.11.943 PMID: 10359546
5. King MC, Marks JH, and Mandell JB, et al (2003) Breast and ovarian cancer risks due to inherited mutations in BRCA1 and BRCA2 Science 302(5645) 643–646 https://doi.org/10.1126/science.1088759 PMID: 14576434
6. Sun J, Meng H, and Yao L, et al (2017) Germline mutations in cancer susceptibility genes in a large series of unselected breast cancer patients Clin Cancer Res 23(20) 6113–6119 https://doi.org/10.1158/1078-0432.CCR-16-3227 PMID: 28724667
7. Petrucelli N, Daly MB, and Pal T (1993) BRCA1- and BRCA2-associated hereditary breast and ovarian cancer GeneReviews® [Internet] eds MP Adam, HH Ardinger, and RA Pagon, et al (Seattle: University of Washington) [http://www.ncbi.nlm.nih.gov/books/NBK1247/] Date accessed: 13/12/20
8. Mittal A and Pramanik R (2021) ASO author reflections: germline testing for all patients with breast cancer: has the time finally come? Ann Surg Oncol 29 1433–1434 https://doi.org/10.1245/s10434-021-10880-8 PMID: 34654988 PMCID: 8519625
9. Yadav S, Hu C, and Hart SN, et al (2020) Evaluation of germline genetic testing criteria in a hospital-based series of women with breast cancer J Clin Oncol 38(13) 1409–1418 https://doi.org/10.1200/JCO.19.02190 PMID: 32125938 PMCID: 7193748
10. Beitsch PD, Whitworth PW, and Hughes K, et al (2019) Underdiagnosis of hereditary breast cancer: are genetic testing guidelines a tool or an obstacle? J Clin Oncol 37(6) 453–460 https://doi.org/10.1200/JCO.18.01631 PMCID: 6380523
11. Yang S, Axilbund JE, and O’Leary E, et al (2018) Underdiagnosis of hereditary breast and ovarian cancer in medicare patients: genetic testing criteria miss the mark Ann Surg Oncol 25(10) 2925–2931 https://doi.org/10.1245/s10434-018-6621-4 PMID: 29998407
12. Mittal A, Deo SVS, and Gogia A, et al (2021) Profile of pathogenic mutations and evaluation of germline genetic testing criteria in consecutive breast cancer patients treated at a North Indian Tertiary Care Center Ann Surg Oncol 29 1423–1432 https://doi.org/10.1245/s10434-021-10870-w PMID: 34601666 PMCID: 8487333
13. Strande NT and Berg JS (2016) Defining the clinical value of a genomic diagnosis in the era of next-generation sequencing Annu Rev Genomics Hum Genet 17 303–332 https://doi.org/10.1146/annurev-genom-083115-022348 PMID: 27362341 PMCID: 7251520
14. Lynce F and Isaacs C (2016) How far do we go with genetic evaluation? Gene, panel, and tumor testing Am Soc Clin Oncol Educ Book 35 e72–e78 https://doi.org/10.1200/EDBK_160391 PMID: 27249773 PMCID: 6054472
15. Catana A, Apostu AP, and Antemie RG (2019) Multi gene panel testing for hereditary breast cancer – is it ready to be used? Med Pharm Rep 92(3) 220–225 PMID: 31460501 PMCID: 6709965
16. Reid S and Pal T (2020) Update on multi-gene panel testing and communication of genetic test results Breast J 26(8) 1513–1519 https://doi.org/10.1111/tbj.13971 PMID: 32639074 PMCID: 7484453
17. Tung N, Domchek SM, and Stadler Z, et al (2016) Counselling framework for moderate-penetrance cancer-susceptibility mutations Nat Rev Clin Oncol 13(9) 581–588 https://doi.org/10.1038/nrclinonc.2016.90 PMID: 27296296 PMCID: 5513673
18. Tischkowitz M, Balmaña J, and Foulkes WD, et al (2021) Management of individuals with germline variants in PALB2: a clinical practice resource of the American College of Medical Genetics and Genomics (ACMG) Genet Med 23(8) 1416–1423 https://doi.org/10.1038/s41436-021-01151-8 PMID: 33976419
19. Moghadasi S, Eccles DM, and Devilee P, et al (2016) Classification and clinical management of variants of uncertain significance in high penetrance cancer predisposition genes Hum Mutat 37(4) 331–336 https://doi.org/10.1002/humu.22956 PMID: 26777316
20. Medendorp NM, Hillen MA, and Murugesu L, et al (2019) Uncertainty related to multigene panel testing for cancer: a qualitative study on counsellors’ and counselees’ views J Community Genet 10(2) 303–312 https://doi.org/10.1007/s12687-018-0393-1 PMCID: 6435776
21. Balmaña J, Digiovanni L, and Gaddam P, et al (2016) Conflicting interpretation of genetic variants and cancer risk by commercial laboratories as assessed by the prospective registry of multiplex testing J Clin Oncol 34(34) 4071–4078 https://doi.org/10.1200/JCO.2016.68.4316 PMID: 27621404 PMCID: 5562435
22. Yohe S and Thyagarajan B (2017) Review of clinical next-generation sequencing Arch Pathol Lab Med 141(11) 1544–1557 https://doi.org/10.5858/arpa.2016-0501-RA PMID: 28782984
23. Easton DF, Pharoah PDP, and Antoniou AC, et al (2015) Gene-panel sequencing and the prediction of breast-cancer risk N Engl J Med 372(23) 2243–2257 https://doi.org/10.1056/NEJMsr1501341 PMID: 26014596 PMCID: 4610139
24. Cline MS, Liao RG, and Parsons MT, et al (2018) BRCA challenge: BRCA exchange as a global resource for variants in BRCA1 and BRCA2 PLoS Genet 14(12) e1007752 https://doi.org/10.1371/journal.pgen.1007752 PMID: 30586411 PMCID: 6324924
25. Spurdle AB, Healey S, and Devereau A, et al (2012) ENIGMA – evidence-based network for the interpretation of germline mutant alleles: an international initiative to evaluate risk and clinical significance associated with sequence variation in BRCA1 and BRCA2 genes Hum Mutat 33(1) 2–7 https://doi.org/10.1002/humu.21628
26. Singh J, Thota N, and Singh S, et al (2018) Screening of over 1000 Indian patients with breast and/or ovarian cancer with a multi-gene panel: prevalence of BRCA1/2 and non-BRCA mutations Breast Cancer Res Treat 170(1) 189–196 https://doi.org/10.1007/s10549-018-4726-x PMID: 29470806
27. Chheda P, Pande S, and Dama T, et al (2020) Spectrum of germline BRCA mutations in hereditary breast and ovarian cancer syndrome in Indian population: a central reference laboratory experience Cancer Res Stat Treat 3(1) 32 https://doi.org/10.4103/CRST.CRST_101_19
28. Kemp Z, Turnbull A, and Yost S, et al (2019) Evaluation of cancer-based criteria for use in mainstream BRCA1 and BRCA2 genetic testing in patients with breast cancer JAMA Netw Open 2(5) e194428 https://doi.org/10.1001/jamanetworkopen.2019.4428 PMID: 31125106 PMCID: 6632150
29. Tung N, Lin NU, and Kidd J, et al (2016) Frequency of germline mutations in 25 cancer susceptibility genes in a sequential series of patients with breast cancer J Clin Oncol 34(13) 1460–1468 https://doi.org/10.1200/JCO.2015.65.0747 PMID: 26976419 PMCID: 4872307
30. Slavin TP, Niell-Swiller M, and Solomon I, et al (2015) Clinical application of multigene panels: challenges of next-generation counseling and cancer risk management Front Oncol 5 208 PMID: 26484312 PMCID: 4586434
31. Chong HK, Wang T, and Lu HM, et al (2014) The validation and clinical implementation of BRCAplus: a comprehensive high-risk breast cancer diagnostic assay PLoS One 9(5) e97408 https://doi.org/10.1371/journal.pone.0097408 PMID: 24830819 PMCID: 4022661
32. Vos J, Gómez-García E, and Oosterwijk JC, et al (2012) Opening the psychological black box in genetic counseling. The psychological impact of DNA testing is predicted by the counselees’ perception, the medical impact by the pathogenic or uninformative BRCA1/2-result Psychooncology 21(1) 29–42 https://doi.org/10.1002/pon.1864
33. Easton DF, Deffenbaugh AM, and Pruss D, et al (2007) A systematic genetic assessment of 1,433 sequence variants of unknown clinical significance in the BRCA1 and BRCA2 breast cancer-predisposition genes Am J Hum Genet 81(5) 873–883 https://doi.org/10.1086/521032 PMID: 17924331 PMCID: 2265654
34. Fackenthal JD, Zhang J, and Zhang B, et al (2012) High prevalence of BRCA1 and BRCA2 mutations in unselected Nigerian breast cancer patients Int J Cancer 131(5) 1114–1123 https://doi.org/10.1002/ijc.27326
35. Kapoor NS, Curcio LD, and Blakemore CA, et al (2015) Multigene panel testing detects equal rates of pathogenic BRCA1/2 mutations and has a higher diagnostic yield compared to limited BRCA1/2 analysis alone in patients at risk for hereditary breast cancer Ann Surg Oncol 22(10) 3282–3288 https://doi.org/10.1245/s10434-015-4754-2 PMID: 26219241
36. Mullai N (2021) The importance of variants of unknown significance (VUS) in BRCA mutation JCO 39(15_suppl) e22507–e22507 https://doi.org/10.1200/JCO.2021.39.15_suppl.e22507
37. Chang J, Seng S, and Yoo J, et al (2019) Clinical management of patients at risk for hereditary breast cancer with variants of uncertain significance in the era of multigene panel testing Ann Surg Oncol 26(10) 3389–3396 https://doi.org/10.1245/s10434-019-07595-2 PMID: 31342386
38. Federici G and Soddu S (2020) Variants of uncertain significance in the era of high-throughput genome sequencing: a lesson from breast and ovary cancers J Exp Clin Cancer Res 39(1) 46 https://doi.org/10.1186/s13046-020-01554-6 PMID: 32127026 PMCID: 7055088
39. Welsh JL, Hoskin TL, and Day CN, et al (2017) Clinical decision-making in patients with variant of uncertain significance in BRCA1 or BRCA2 Genes Ann Surg Oncol 24(10) 3067–3072 https://doi.org/10.1245/s10434-017-5959-3 PMID: 28766224 PMCID: 6171336
40. Slavin TP, Van Tongeren LR, and Behrendt CE, et al (2018) Prospective study of cancer genetic variants: variation in rate of reclassification by ancestry J Natl Cancer Inst 110(10) 1059–1066 https://doi.org/10.1093/jnci/djy027 PMID: 29618041 PMCID: 6249694
Supplementary Material
Supplementary Table 1. NCCN checklist.

Supplementary Table 2. MCG plus checklist.
Supplementary Table 3. Details of VUS in all study population.