Proteomics characterisation of central nervous system metastasis biomarkers in triple negative breast cancer
Katerin Rojas L1*, Lucía Trilla-Fuertes2*, Angelo Gámez-Pozo2,3, Cristina Chiva4,5, Juan Sepúlveda1, Luis Manso1, Guillermo Prado-Vázquez2,3, Andrea Zapater-Moros2,3, Rocío López-Vacas3, María Ferrer-Gómez3, César Mendiola1, Enrique Espinosa6,7, Eduard Sabidó4,5, Eva Ciruelos1 and Juan Ángel Fresno Vara2,3,7
1Department of Medical Oncology, Hospital Universitario 12 de Octubre, 28041 Madrid, Spain
2Biomedica Molecular Medicine SL, 28049 Madrid, Spain
3Molecular Oncology and Pathology Lab, Instituto de Genética Médica y Molecular-INGEMM, Hospital Universitario La Paz-IdiPAZ, 28029 Madrid, Spain
4Proteomics Unit, Center of Genomics Regulation, Barcelona Institute of Science and Technology, 08036 Barcelona, Spain
5Proteomics Unit, Universitat Pompeu Fabra, 08002 Barcelona, Spain
6Medical Oncology Service, Hospital Universitario La Paz-IdiPAZ, 28029 Madrid, Spain
7CIBERONC, Instituto de Salud Carlos III, 28029 Madrid, Spain
*Katerin L Rojas and Lucía Trilla-Fuertes contributed equally to this work.
Correspondence to: Juan Ángel Fresno Vara. Email: juanangel.fresno@salud.madrid.org
Abstract
Background: Breast cancer (BC) is the most frequent tumour in women. Triple negative tumours (TNBC)–which are associated with minor survival rates—lack markers predictive of response to anticancer drugs. Triple negative tumours frequently metastasise to the central nervous system (CNS).
Objective: The main objective of this study was to study differences in tumour protein expression between patients with CNS metastases and those without this kind of spread, and propose new biomarkers.
Methods: A retrospective study was performed. Targeted proteomics and statistical analyses were used to identify possible biomarkers.
Results: Proteins were quantified by a targeted proteomics approach and protein expression data were successfully obtained from 51 triple negative formalin-fixed paraffin-embedded samples. ISG15, THBS1 and AP1M1 were identified as possible biomarkers related with CNS metastasis development.
Conclusions: Three possible biomarkers associated with CNS metastases in TNBC tumours were identified: ISG15, THBS1 and AP1M1. They may become markers predicting the appearance of CNS infiltration in triple negative BC.
Keywords: breast cancer, triple negative, proteomics, central nervous system metastases, biomarkers, personalised medicine
Copyright: © the authors; licensee ecancermedicalscience. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/3.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Published: 15/01/2019; Received: 12/09/2018
Introduction
Triple-negative breast cancer (TNBC) subtype represents approximately 10%–20% of all cases of breast cancer (BC) in Caucasian women and is associated with poor prognosis in terms of distant relapse-free survival and overall survival (OS) [1–3]. Patients with TNBC do not benefit from targeted therapies because a therapeutic target has not yet been established [1–3].
In murine models of BC, cyclooxygenase COX-2, an epidermal growth factor receptor (EGFR) ligand and the ST6GALNAC5 gene were identified as candidate genes related to the development of brain metastasis [4]. We have previously analysed the expression of these three genes in formalin-fixed paraffin-embedded (FFPE) samples from patients with TNBC and central nervous system (CNS) involvement and did not find such a correlation [5].
Mass-spectrometry-based proteomics is beginning to develop and to mature through a blend of enhanced instrumentation, sample preparation strategies and computational investigation [6–8]. These advances permit the distinguishing proof of thousands of proteins from tissue compatible with clinical daily practice, which is pertinent for the investigation of complex ailments. Proteomics investigations are important to describe the entire situation of flagging pathways and modified natural procedures because of the particular change set in every tumour, so protein profiling harbours the possibility of building up new patient stratifications and biomarkers advancing in individualised treatment [9].
Besides shotgun proteomics approaches, targeted proteomics procedures, such as parallel reaction monitoring (PRM) [10, 11], allow the measurement with high exactness and precision of a set of pre-selected set of proteins of interest. This technique was previously used by our group to validate predictive signatures in TNBC patients treated with adjuvant chemotherapy [8].
In this study, we aimed to identify proteins predicting CNS metastases and therapeutic target candidates in patients with TNBC. We defined three candidate biomarkers related to the presence of CNS metastasis using targeted proteomics techniques.
Brain metastases generally tend to occur late in the course of metastatic breast cancer and are typically associated with 1 and 2 year survival rates of only 20% and < 2%, respectively. In addition, several studies have reported a greater propensity of ER-negative tumours metastasizing to the brain compared with ER-positive tumors [29, 30]. Molecular detailing of subsequent events of penetration, seeding, and outgrowth in the brain is highly relevant for developing therapeutic strategies to treat or prevent, CNS metastases.
Material and methods
Samples
Fifty-one TNBC samples from primary tumours from I+12 Biobank (RD09/0076/00118) integrated at the Spanish Hospital Biobank Network (RetBioH; www.redbiobancos.es) were included in the study. All patients were treated with adjuvant chemotherapy. The histopathological features and tumour content of each sample were confirmed by an experienced pathologist. Eligible samples had to include at least 50% of tumour cells. Informed consent from all individual participants included in the study and approval from the Ethical Committee of Hospital Universitario La Paz (HULP PI-1417) were obtained for the conduct of the study. Samples from this cohort were analysed in previous works [8].
Total protein extraction
Proteins were extracted from FFPE samples as described in previous works [12]. Briefly, protein extracts were prepared in 2% sodium dodecyl sulfate (SDS) following a heat-induced antigen retrieval protocol [13]. Then, protein extracts were digested with trypsin and SDS was removed using Detergent Removal Spin Columns (Pierce).
Parallel reaction monitoring data acquisition and analysis
As described in detail previously, 37 proteins were selected for performing the PRM experiments based on their prognostic value in TNBC [8]. One to four unique peptides per protein were selected for quantification by PRM, as described in previous studies [8]. Briefly, selected peptides were spiked in the peptide mixture as isotopically labelled internal standard peptides. Each sample was analysed using an Orbitrap Fusion Lumos (Thermo Fisher Scientific) coupled to an EASY-nanoLC 1000 Ultra Performance Liquid Chromatography (UPLC) system (Thermo Fisher Scientific). A previously described scheduled PRM method was used for data acquisition [8]. Mass-spectrometry (MS) fragmentation was performed as described before [8]. The Parallel Reaction Monitoring dataset is publicly available in the Panorama web server at https://panoramaweb.org/labkey/QrYeZ2.url. Product ion chromatographic traces corresponding to the targeted precursor peptides were evaluated with Skyline software v 2.5 [8]. For each monitored peptide, a light-to-heavy ratio was calculated per patient. Ratios were transformed to the logarithmic scale (log2) and the obtained values were used as a proxy for the protein amount.
Statistical analyses
Statistical analyses were done using GraphPad Prism v 6. Class comparison analyses were performed in BRB Array Tools developed by Dr Richard Simons. P-values are two-sided and p <0.05 are considered statistically significant.
Results
Patients’ characteristics
Fifty-one samples characterised as TNBC tumours were included. Table 1 shows the patients’ clinical features. The median follow-up was 42 months (range 1–236 months). Twenty-two patients developed CNS metastases. Patients who developed CNS metastasis have a poor distant metastasis-free survival (DMFS) and OS (Figures 1 and 2).
Targeted proteomics
Thirty-seven proteins selected for their prognostic value in TNBC tumours based on results from previous works [8] were measured using PRM targeted proteomics. One to four unique peptides per protein were chosen and these peptides were used to perform a class comparison analysis.
Differential protein expression between CNS and no CNS TNBC metastatic tumours
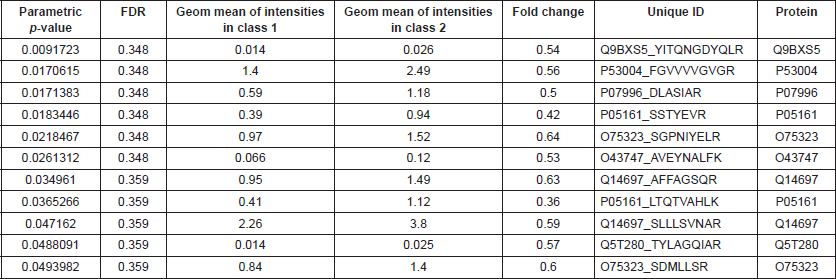
BRB Array Tool was employed to establish protein candidates differentially expressed between the two categories. Class comparison analysis, based on a parametric t-test, and volcano plot showed 11 peptides with a significant differential expression between no CNS metastasis and CNS metastasis patients (Table 2, Figure 2) (p-value = 0.05). However, a Mann–Whitney test (proteomics data never follow a normal distribution) reduced these 11 proteins into four candidate peptides, two of them belonging to the same protein: P05161 (interferon simulated gene 15 ubiquitin-like modifier, ISG15), Q9BXS5 (AP-1 complex subunit μ−1, AP1M1) and P07996 (thrombospondin-1, THBS1) (Figure 3). All of these proteins are overexpressed in TNBC tumours which develop CNS metastases comparing with TNBC tumours which do not develop them.
Discussion
Proteomics technologies have been used to uncover biomarkers and molecular mechanisms associated with BC [14]. TNBC tumours frequently metastasise to the lungs and the brain [8, 12, 13]. In this work, using targeted proteomics and statistical analyses, we characterised protein expression patterns in tumours with and without CNS spread.
A group of three genes had previously been related to the presence of brain metastases in BC in murine models [4] but we failed to validate them in clinical samples [5]. Differential protein expression between the primary tumour and the brain metastasis could explain this result. We then decided to use PRM and class comparison analysis as an alternative and more powerful approach.
Table 1. Patients’ characteristics.
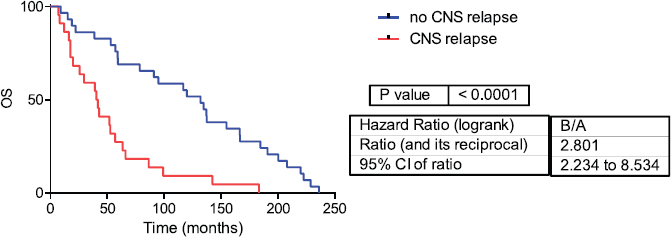
Figure 1: Overall survival in CNS and no CNS relapse tumours. OS= Overall survival.
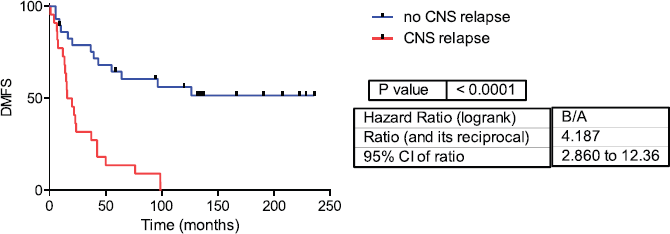
Figure 2: Distant metastasis-free survival in CNS and no CNS relapse tumours. DMFS= Distant metastasis-free survival.
Table 2. Differential peptides between CNS and no CNS metastatic patients.
ISG15, an interferon (IFN)-inducible, ubiquitin-like protein, was overexpressed in TNBC tumours that developed CNS metastases. ISG15 protein is involved in numerous cellular functions, including the interferon-induced immune response and the regulation of cellular protein turnover [15, 16]. Desai et al. [17] demonstrated that free ISG15 and its protein conjugated form (ISGylation) are increased in human solid tumours and tumour cell lines compared with their respective normal counterparts. Other study identified in BC tumours elevated ISG15 expression when compared with normal tissue [18]. This overexpression was independent of HER2, progesterone receptor and oestrogen receptor status and correlated with an unfavourable prognosis and poor response to chemotherapy and radiation [19]. Additionally, a relationship between this protein and motility in BC tumours has been previously described [20]. ISG15 was also previously proposed as a biomarker with prognostic significance in BC; however, this study did not differentiate between BC subtypes [18].
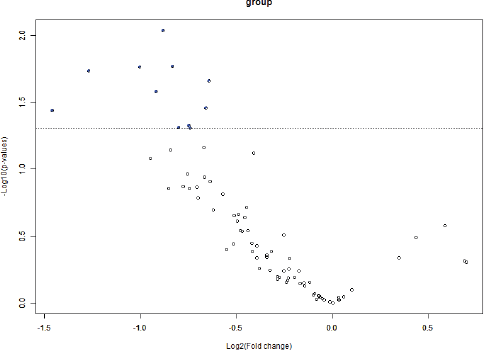
Figure 3: Volcano plot showing differential peptides between tumors with presence and absence of CNS metastases. Differential peptides, which are colored in blue, between CNS and no CNS metastatic tumors (p<0.05).
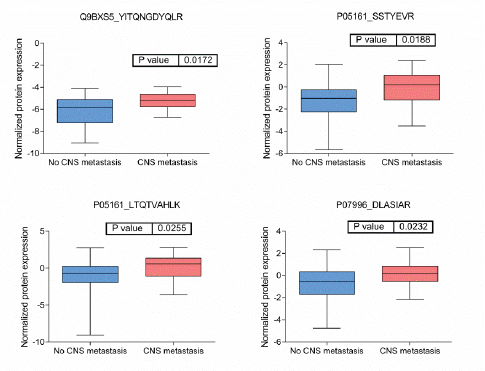
Figure 4: Boxplots of relative protein expression of differential peptides between tumors with no and with CNS metastases. Relative protein expression of peptides which presented a differential expression between tumors with central nervous system metastasis and tumors which no developed central nervous system metastasis.
THBS1, thrombospondin1, is an adhesive glycoprotein belonging to thrombospondins family that mediates matrix interactions and extracellular matrix structure [21]. It was overexpressed in our TNBC tumour samples which developed CNS metastases. THBS1 is an angiogenesis inhibitor [22] and it is overexpressed in BC plasma samples compared with normal samples, suggesting this molecule as a good serological marker [23]. In addition, previous studies established that THBS1 promotes metastasis in murine BC models [24]. This protein was also included in a prognostic signature for TNBC recurrence and proposed as a bad prognostic biomarker [25].
AP1M1, adaptor-related protein complex 1 μ 1 subunit, is the medium chain of the clathrin-associated protein complex AP-1. AP-1 complex is located at the Golgi and it is implicated on endocytosis [26]. It was also overexpressed in TNBC tumours with CNS metastases. It was described that AP-1 is necessary for some antigen presentation processes by major histocompatibility complex (MHC) molecules [27]. In addition, this complex was previously associated with metastasis using murine models of epidermal tumours.
Our study has some limitations. Although it is necessary to validate these findings in a new cohort, these proteins may represent novel TNBC tumour markers helpful in selecting patients who will develop CNS metastases. Although it may be interesting to correlate proteomics findings with mutational data, at this moment, Next-Generation Sequencing data from these patients are not available. They should also be explored as therapeutic targets in this clinical context. On the other hand, proteomics currently provides a powerful tool for basic research, clinical diagnostics and drug development applications in combination with advanced bioinformatics and large databases. However, an improvement in data acquisition and data analysis in targeted proteomics techniques is still necessary.
Conclusions
Current efforts to treat CNS metastases in TNBC tumours are limited, and drugs used have proven effects on the primary tumours but lack specificity for the intracerebral tumours, passing the blood-brain barrier and intracerebral tumour cell growth. The identification of biomarkers for CNS metastases in TNBC are not well established. In this study, we proposed three new proteins related with CNS metastases in TNBC tumours. As far as we know, this is the largest cohort with CNS metastasis data analysed by proteomics. The clinical value of ISG15, AP1M1 and THBS1 as either diagnostic/prognostic factors or as therapeutic targets must be validated in independent cohorts.
Funding
This study was funded by Instituto de Salud Carlos III, Spanish Economy and Competitiveness Ministry, Spain and by the FEDER programme, ‘Una forma de hacer Europa’ (PI12/01016, PI12/00444 and PI15/01310). LT-F is supported by the Spanish Economy and Competitiveness Ministry (DI-15-07614). GP-V is supported by Consejería de Educación, Juventud y Deporte of Comunidad de Madrid (IND2017/BMD7783). The Centre of Genomics Regulation/Universitat Pompeu Fabra Proteomics Unit is part of the ‘Plataforma de Recursos Biomoleculares y Bioinformáticos (ProteoRed)’ supported by a grant of Instituto de Salud Carlos III and Spanish Ministry of Economy and Competitiveness (PT13/0001). We acknowledge the support of the Spanish Ministry of Economy and Competitiveness, ‘Centro de Excelencia Severo Ochoa 2013–2017’ (SEV-2012-0208) and from ‘Secretaria d’Universitats i Recerca del Departament d’Economia i Coneixement de la Generalitat de Catalunya’ (2014SGR678).
Conflicts of interest
JAFV, EE and AG-P are shareholders in Biomedica Molecular Medicine SL. LT-F is an employee of Biomedica Molecular Medicine SL. The other authors declare no competing interests.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. For this type of study, formal consent is not required.
References
1. Lehmann BD, Bauer JA, and Chen X, et al (2011) Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies J Clin Invest 121(7) 2750–2767 https://doi.org/10.1172/JCI45014 PMID: 21633166 PMCID: 3127435
2. Lachapelle J and Foulkes WD (2011) Triple-negative and basal-like breast cancer: implications for oncologists Curr Oncol 18(4) 161–164 https://doi.org/10.3747/co.v18i4.824 PMID: 21874112 PMCID: 3149541
3. Heitz F, Harter P, and Lueck HJ, et al (2009) Triple-negative and HER2-overexpressing breast cancers exhibit an elevated risk and an earlier occurrence of cerebral metastases Eur J Cancer 45(16) 2792–2798 https://doi.org/10.1016/j.ejca.2009.06.027 PMID: 19643597
4. Bos PD, Zhang XH, and Nadal C, et al (2009) Genes that mediate breast cancer metastasis to the brain Nature 459(7249) 1005–1009 https://doi.org/10.1038/nature08021 PMID: 19421193 PMCID: 2698953
5. Laimito KR, Gámez-Pozo A, and Sepúlveda J, et al (2016) Characterisation of the triple negative breast cancer phenotype associated with the development of central nervous system metastases Ecancermedicalscience 10 632 PMID: 27170832 PMCID: 4854224
6. Gámez-Pozo A, Berges-Soria J, and Arevalillo JM, et al (2015) Combined label-free quantitative proteomics and microRNA expression analysis of breast cancer unravel molecular differences with clinical implications Cancer Res 2243–2253 75(11) https://doi.org/10.1158/0008-5472.CAN-14-1937 PMID: 25883093
7. Gámez-Pozo A, Trilla-Fuertes L, and Berges-Soria J, et al (2017) Functional proteomics outlines the complexity of breast cancer molecular subtypes Scientific Reports 7(1) 10100 https://doi.org/10.1038/s41598-017-10493-w PMID: 28855612 PMCID: 5577137
8. Gámez-Pozo A, Trilla-Fuertes L, and Prado-Vázquez G, et al (2017) Prediction of adjuvant chemotherapy response in triple negative breast cancer with discovery and targeted proteomics PLoS One 12(6) e0178296 https://doi.org/10.1371/journal.pone.0178296 PMID: 28594844 PMCID: 5464546
9. Bernhardt S, Bayerlová M, and Vetter M, et al (2017) Proteomic profiling of breast cancer metabolism identifies SHMT2 and ASCT2 as prognostic factors Breast Cancer Res 19(1) 112 https://doi.org/10.1186/s13058-017-0905-7 PMID: 29020998 PMCID: 5637318
10. Peterson AC, Russell JD, and Bailey DJ, et al (2012) Parallel reaction monitoring for high resolution and high mass accuracy quantitative, targeted proteomics Mol Cell Proteomics 11(11) 1475–1488 https://doi.org/10.1074/mcp.O112.020131 PMID: 22865924 PMCID: 3494192
11. Gallien S, Kim SY, and Domon B (2015) Large-Scale targeted proteomics using internal standard triggered-parallel reaction monitoring (IS-PRM) Mol Cell Proteomics 14(6) 1630–1644 https://doi.org/10.1074/mcp.O114.043968 PMID: 25755295 PMCID: 4458725
12. Gámez-Pozo A, Ferrer NI, and Ciruelos E, et al (2013) Shotgun proteomics of archival triple-negative breast cancer samples Proteomics Clin Appl 7(3–4) 283–291 https://doi.org/10.1002/prca.201200048 PMID: 23436753
13. Gámez-Pozo A, Sánchez-Navarro I, and Calvo E, et al (2011) Protein phosphorylation analysis in archival clinical cancer samples by shotgun and targeted proteomics approaches Mol Biosyst 7(8) 2368–2374 https://doi.org/10.1039/c1mb05113j PMID: 21617801
14. Vitzthum F, Behrens F, and Anderson NL, et al (2005) Proteomics: from basic research to diagnostic application. A review of requirements and needs J Proteome Res 4(4) 1086–1097 https://doi.org/10.1021/pr050080b PMID: 16083257
15. Owhashi M, Taoka Y, and Ishii K, et al (2003) Identification of a ubiquitin family protein as a novel neutrophil chemotactic factor Biochem Biophys Res Commun 309(3) 533–539 https://doi.org/10.1016/j.bbrc.2003.08.038 PMID: 12963022
16. Desai SD, Haas AL, and Wood LM, et al (2006) Elevated expression of ISG15 in tumor cells interferes with the ubiquitin/26S proteasome pathway Cancer Res 66(2) 921–928 https://doi.org/10.1158/0008-5472.CAN-05-1123 PMID: 16424026
17. Desai SD (2015) ISG15: A double edged sword in cancer Oncoimmunology 4(12) e1052935 https://doi.org/10.1080/2162402X.2015.1052935 PMID: 26587329 PMCID: 4635879
18. Bektas N, Noetzel E, and Veeck J, et al (2008) The ubiquitin-like molecule interferon-stimulated gene 15 (ISG15) is a potential prognostic marker in human breast cancer Breast Cancer Res 10(4) R58 https://doi.org/10.1186/bcr2117 PMID: 18627608 PMCID: 2575531
19. Weichselbaum RR, Ishwaran H, and Yoon T, et al (2008) An interferon-related gene signature for DNA damage resistance is a predictive marker for chemotherapy and radiation for breast cancer Proc Natl Acad Sci USA 105(47) 18490–18495 https://doi.org/10.1073/pnas.0809242105 PMID: 19001271 PMCID: 2587578
20. Desai SD, Reed RE, and Burks J, et al (2012) ISG15 disrupts cytoskeletal architecture and promotes motility in human breast cancer cells Exp Biol Med (Maywood) 237(1) 38–49 https://doi.org/10.1258/ebm.2011.011236
21. Chen H, Herndon ME, and Lawler J (2000) The cell biology of thrombospondin-1 Matrix Biol 19(7) 597–614 https://doi.org/10.1016/S0945-053X(00)00107-4 PMID: 11102749
22. Lawler PR and Lawler J (2012) Molecular basis for the regulation of angiogenesis by thrombospondin-1 and -2 Cold Spring Harb Perspect Med 2(5) a006627 https://doi.org/10.1101/cshperspect.a006627 PMID: 22553494 PMCID: 3331684
23. Suh EJ, Kabir MH, and Kang UB, et al (2012) Comparative profiling of plasma proteome from breast cancer patients reveals thrombospondin-1 and BRWD3 as serological biomarkers Exp Mol Med 44(1) 36–44 https://doi.org/10.3858/emm.2012.44.1.003 PMCID: 3277896
24. Yee KO, Connolly CM, and Duquette M, et al (2009) The effect of thrombospondin-1 on breast cancer metastasis Breast Cancer Res Treat 114(1) 85–96 https://doi.org/10.1007/s10549-008-9992-6 PMCID: 2631620
25. Campone M, Valo I, and Jézéquel P, et al (2015) Prediction of recurrence and survival for triple-negative breast cancer (TNBC) by a protein signature in tissue samples Mol Cell Proteomics 14(11) 2936–2946 https://doi.org/10.1074/mcp.M115.048967 PMID: 26209610 PMCID: 4638037
26. Hirst J and Robinson MS (1998) Clathrin and adaptors Biochim Biophys Acta 1404(1–2) 173–193 https://doi.org/10.1016/S0167-4889(98)00056-1 PMID: 9714795
27. Kulpa DA, Del Cid N, and Peterson KA, et al (2013) Adaptor protein 1 promotes cross-presentation through the same tyrosine signal in major histocompatibility complex class I as that targeted by HIV-1 J Virol 87(14) 8085–8098 https://doi.org/10.1128/JVI.00701-13 PMID: 23678182 PMCID: 3700188
28. Miliani de Marval P, Lutfeali S, and Jin JY, et al (2011) CYLD inhibits tumorigenesis and metastasis by blocking JNK/AP1 signaling at multiple levels Cancer Prev Res (Phila) 4(6) 851–859 https://doi.org/10.1158/1940-6207.CAPR-10-0360
29. Tham YL, Sexton K, and Kramer R et al (2006) Primary breast cancer phenotypes associated with propensity for central nervous system metastases Cancer 107 696-704
30. Samaan NA, Buzdar AU, and Aldinger KA et al (1981) Estrogen receptor: a prognostic factor in breast cancer Cancer 47 554-560