Mutational frequency of KRAS, NRAS, IDH2, PIK3CA, and EGFR in North Indian gallbladder cancer patients
Aarti Sharma1, Ashok Kumar1, Niraj Kumari2, Narendra Krishnani2, and Neeraj Rastogi3
1Department of Surgical Gastroenterology, Sanjay Gandhi Post Graduate Institute of Medical Sciences (SGPGIMS), Lucknow, Uttar Pradesh, 226014, India
2Department of Pathology, Sanjay Gandhi Post Graduate Institute of Medical Sciences (SGPGIMS), Lucknow, Uttar Pradesh, 226014, India
3Department of Radiotherapy, Sanjay Gandhi Post Graduate Institute of Medical Sciences (SGPGIMS), Lucknow, Uttar Pradesh, 226014, India
Correspondence to: Ashok Kumar. Email: doc.ashokgupta@gmail.com and akgupta@sgpgi.ac.in
Abstract
Background: Gallbladder cancer (GBC) has a peculiar geographical distinction, with a high prevalence seen in North India and Chile. There are various aetiopathogenetic mechanisms of GBC causation; one of them is a series of pathogenic mutations, which is responsible for the malignant transformation of gallbladder epithelium. Therefore, the present study aimed to find out cancer-specific hot spot mutations in five major cancer-related genes KRAS exon1 &2, NRAS exon1, IDH2 exon, PIK3CA exon 20, IDH2 exon 4 and EGFR exon 20 in North Indian GBC patients and their association with clinicopathological variables.
Material and methods: This study included 34 histopathologically confirmed GBC cases. The clinical material consisted of formalin-fixed paraffin-embedded (FFPE) blocks of the patients. DNA isolation was done from FFPE tissue. DNA sequencing was performed by the capillary electrophoresis method. The chi-square (χ2) test was used to test for a statistically significant relationship between two categorical study variables.
Results: The overall incidence of somatic mutations in KRAS exon 1&2, NRAS exon1, IDH2 exon4, PIK3CA exon20, and EGFR exon 20 in Indian GBC patients was found in 8/34 (23.5%), 3/34 (8.8%), 4/34 (11.7%), 7/34 (20.6%), 7/34 (20.6%), respectively. KRAS exon 1 and two mutations were found to be significantly associated with advanced stage GBC patients.
Conclusion: KRAS, PIK3CA, and EGFR were found to be the most frequently mutated genes among the five tested in this study.
Keywords: gallbladder cancer (GBC), formalin-fixed paraffin-embedded (FFPE), genetic mutations.
Copyright: © the authors; licensee ecancermedicalscience. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (
Published: 07/08/2017; Received: 08/02/2017
Introduction
Gallbladder cancer (GBC) has a peculiar geographical distinction, with a high prevalence observed in North India and Chile [1]. However, its frequency in western countries is relatively less [2]. GBC accounts only for 0.5% of all gastrointestinal malignancies cancers in the United States [3]. The 2000–2013 figures from the Surveillance, Epidemiology, and End Results (SEER 18) show that age-adjusted incidence rate of GBC in 2013 is 1.1528 per 100,000 in the Asian population https://seer.cancer.gov/. Surgical resection remains the only chance of cure, but it is possible in only a small percentage of patients with GBC who are diagnosed early. The overall five-year survival rate for GBC is 32% but for advanced stage, it declines to 10% [4, 5].
There are many pathogenesis means that have been proposed for the occurrence of GBC, which include chronic inflammation, environmental factors, dietary changes, and genetic factors. The less understood aspect for gallbladder pathogenesis is genetic alteration. From the earliest observation that abnormal gene expression could result from a single mutation that can drive a series of events and finally leading to malignant transformation, the mutation continues to evolve as a central mechanism in cancer biology.
The role of various genetic mutations in GBC is a field of active research nowadays. Somatic mutations occurring in intracellular signalling pathways cause aberrant activation of the signalling molecules. These mutations have transformed the diagnosis and treatment in some cancers [6–8]. Some of these genetic changes are associated with particular risk factors, whereas some changes are associated with differences in prognosis [9]. Different subtypes of cancer harbour specific gene mutations that act as invaluable markers for disease diagnosis and prognosis, for example, the leukaemia cells of patients with chronic myeloid leukaemia (CML) contain a mutated gene called BCR-ABL.
The identification of key genetic mutations in GBC that could be involved in carcinogenesis may help to better understand the molecular basis of pathogenesis and possibly help in improving treatment strategies. The mutations of RAS (KRAS and NRAS), IDH2, EGFR, and PI3K are clinically relevant and well associated in many other cancers [10–15]. Therefore, the present study aimed to find out cancer-specific hot spot mutations in five major cancer-related genes KRAS exon1 &2, NRAS exon 1, PIK3CA exon 20, IDH2 exon 4, and EGFR exon 20 in North Indian GBC patients and their association with clinicopathological variables through a hospital-based study.
Methodology
Sample collection and DNA isolation
In this study, we have included 34 histopathologically confirmed GBC cases, which were treated in the Department of Surgical Gastroenterology and Radiotherapy. Formalin-fixed paraffin-embedded blocks of the patients, stored as archival material in Department of Pathology were taken as clinical material, without any compromise with histopathological quality. Tumour area was identified by staining a three micron thick section with haematoxylin and eosin, and then, the slides were visualised microscopically. The tumour area having greater than 80% tumour cells was marked. For DNA isolation, we cut some other 20 micron thick sections from the same block using microtome and collected on a glass slide. The slide was fixed on a hot plate at 65° for one to two hours. These sections were then deparaffinised with two washes of xylene followed by rehydration with a graded alcohol (30%, 50%, 70%, 100%) washes. The tumorous area was scarped using a sharp scalpel. DNA extraction was done using commercially available Quigen FFPE DNA extraction kit (Qiagen, Valencia, CA) as per manufacture instructions.
DNA quality control
DNA concentration was determined using spectrophotometer (NanodropTM, Thermo Scientific, USA). DNA having minimum concentration of 200 ng/μl was considered for further analysis.
Primer designing
For primer designing we used primer3 software (http://primer3.sourceforge.net/). The desired sequence of the gene of interest was taken from the NCBI database against which the software gave four sets of primers. The primer pair < 200 bp in length, with annealing temperature between 48–58°, and GC content not exceeding 60% was selected for the polymerase chain reaction (PCR). All primers were binding at a site 70–80 bp away from mutation of interest so interpretation of mutational region was proper and free from noise. The primer sequences used for sequencing analysis are given in Table 1.
DNA sequencing
PCR amplification products generated by the Lightcycler PCR were purified using QiaQuick reagents (Qiagen, Valencia, CA) and were cycle sequenced using Big Dye v3.1 reagents (Applied Biosystems, Weiterstadt, Germany) according to the manufacturer’s protocol. Sequencing products were purified with CleanSEQ Sequencing Purification System (Agencourt Bioscience Corp., Beverly, MA) and automated sequencing performed by capillary electrophoresis on an ABI3700 (Applied Biosystems). Sequences were aligned and examined by two separate approaches: electronically with a set threshold of 10% and by visual inspection of the electropherogram, using Finch TV software (
Statistical and in silico analysis
All the data obtained by the experiments also with clinical details were analysed using SPSS version 16.0 (SPSS, USA). Descriptive statistics of study subjects are either represented as mean with standard deviation or frequency with percentages. The chi-square (χ2) test was used to test for a statistically significant relationship between two categorical study variables. A cut-off of 0.05 for p values was used as a limit of significance to predict the associations. In addition, the functional consequences of studied mutations were enquired from online webserver COSMIC (http://cancer.sanger.ac.uk/cosmic) that uses FATHMM algorithm (Functional analysis through hidden Markov Models) to predict the mutation impact. Any scores above ≥ 0.7 were classified as ‘Pathogenic’.
Table 1. Primer sequences for PCR.

Results
Characteristic profile of the study subjects
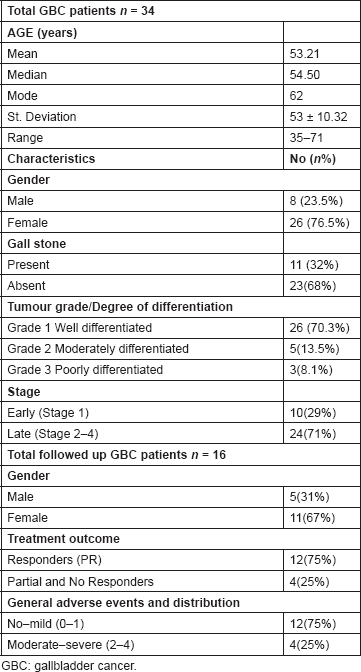
This study included 34 GBC cases which included eight (23.5%) males and 26 (76.5%) females with the mean age of 53.21 ± 10.32 (range 35–71 years). Follow-up data were available for 16 patients only who continued their post-surgery treatment in the Department of Radiotherapy. Follow-up data were collected every three months from the time of enrolment until death or last scheduled follow-up. Characteristics of GBC patients along with all the details regarding treatment response, drug-related toxicity, tumour grade, etc. are listed in Table 2.
Table 2. Characteristics of GBC patients.
Mutations in KRAS oncogene
KRAS (Kirsten ras oncogene homolog) gene belongs to mammalian ras gene family. Any single-nucleotide change can lead to substitution of an amino acid, which finally is responsible for an activating mutation.
We evaluated a total of eight mutations present in exons 1 and exon 2 in KRAS oncogene in 34 GBC by performing PCR and DNA sequencing. KRAS mutations were determined in 8/34 (23.5%) cases; most of the patients have exon 1 mutations, only two cases have presence of exon 2 mutations (Table 3).
Mutations in NRAS oncogene
N-RAS (neuroblastoma RAS viral oncogene homologue) is a proto-oncogene which encodes a membrane protein that acts as a shuttle between the Golgi apparatus and the plasma membrane. Mutations in this gene have been seen to be associated with rectal cancer, follicular thyroid cancer, etc. [11, 12].
Table 3. Overview of gene mutations found in GBC patients.
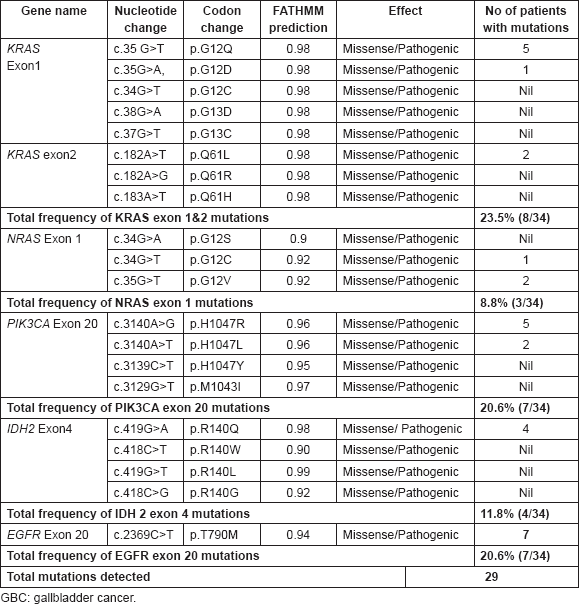
We sequenced the exon 2 of NRAS gene to look at the hotspot mutations, that is, p.G12S, p.G12C and p.G12V, which have already been reported as pathogenic in cosmic database. The variants of p.G12V and p.G12C as two hotspot mutations of the NRAS exon 1 were found only in 3 out of 34 tumours. No p.G12S, mutation was seen in any of the GBC patients Table 3.
Mutation in PIK3CA gene
PIK3CA (phosphatidylinositol 3-kinase) gene plays a key role in cell growth, survival, proliferation, motility, and morphology. This gene has also been reported as oncogenic and involved in cervical cancer [13].
Here, we have sequenced exon 20 of PIK3CA gene. It was found that PIK3CA p.H1047R(c.3140A>G) mutation was present in 5/34 GBC patients. Whereas two cases were harbouring p.H1047L(c.3140A>T) mutation (Table 3).
Mutation in IDH2 gene
Isocitrate dehydrogenase gene codes for a digestive enzyme that catalyses the oxidative decarboxylation of isocitrate to 2-oxoglutarate in the citric acid cycle. This gene is associated with glial tumours [14].
We sequenced exon 4 Of IDH2 gene to look for the pathogenic mutations that are already reported in cosmic database in GBC patients. Only four cases were positive for p.R140Q c.419G>A mutations. Other pathogenic mutations of exon 4, that is, p.R140W, c.418C>T p.R140L, c.419G>T, p.R140G c.418C>G were absent in all studied GBC cases (Table 3).
Mutation in EGFR gene
EGFR is a cell surface protein that codes for epidermal growth factor receptor protein. It has a major role in cell proliferation. Mutations in this gene are found to be associated with Lung cancer [15].
We characterised one non-synonymous, pathogenic mutation p.T790M c.2369C>T in exon 20 of 7/34 GBC patients (Table 3).
Out of all studied mutations, the most common somatic mutations detected were KRAS exon 1 and 2 in 8/34 (23.6%) cases followed by PIK3CA exon 20 in 7/34 (20.6%) and EGFR exon 20 in 7/34 (20.6%) cases. NRAS exon 1 and IDH2 exon 4 were found mutated in least number of GBC cases (8.8% and 11.8%, respectively) (all data shown in Table 3).
Association of gene mutations with clinicopathological data
We examined the relation between gene mutation status and various clinicopathological features, including age at diagnosis, degree of differentiation, stage of tumour and gender. There was no significant association of any of the studied mutations with clinicopathological features except that KRAS exon 1 and 2 mutations were found to be significantly associated with advanced stage GBC patients (Table 4).
Discussion
Occurrence of mutations can be responsible for the malignant transformation of gallbladder epithelium. The tumour causing genes can belong to either tumour suppressor or oncogenes or even DNA repair genes. Any error in any one of the genes can become mutations, which may eventually lead to cancer. GBC also involves the changes in multiple oncogenes and tumour suppressor genes. In future, many of these mutated genes can also represent targets for novel therapeutic agents that are more specific, more efficacious, and less toxic than broad-based chemotherapeutic regimens.
Table 4. Relationship of detected mutations with clinicopathological features in GBC patients.
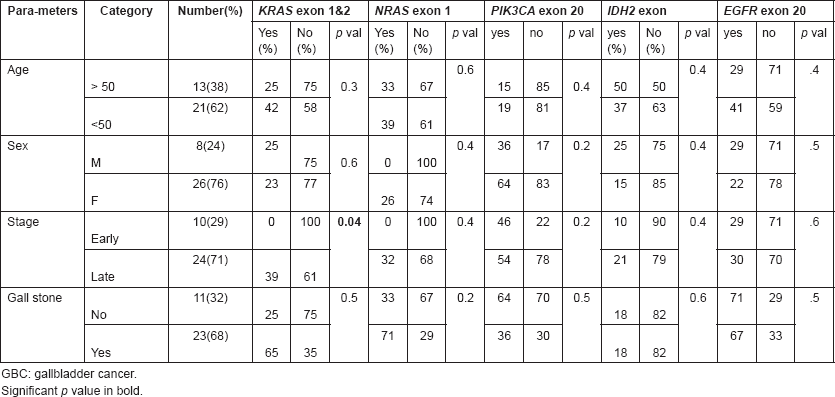
In the present series, we have studied the frequency of pathogenic mutations in five major cancer-related genes in north Indian GBC patients. Population-based data show that North India has high prevalence of GBC [1]. To study the mutational frequency of most common cancer-related genes, we performed Sanger sequencing of 34 GBC patients. The overall incidence of somatic mutations in KRAS exon 1 and 2, NRAS exon 1, IDH2 exon 4, PIK3CA exon 20, and EGFR exon 20 in Indian GBC patients was found to be 8/34 (23.5%), 3/34 (8.8%), 4/34 (11.7%), 7/34 (20.6%), 7/34 (20.6%), respectively. Though most of the mutations were not linked to stage or grade of the disease, KRAS exon 1 and 2 mutations were found mainly in cases with stage 3 and 4.
A genetic model for gallbladder carcinogenesis by S. G Barreto et al has also shown the presence of KRAS mutations as an event of GBC progression to advanced stage [16]. However, many other genetic alterations observed not only in advanced cancers but also in pre-neoplastic lesions depends on type and severity of the tumour [17]. The presence of loss of heterozygosity and TP53 mutations are the most frequent and early events in GBC (~50%) and they are observed even in normal-like tissues exposed to gallstones [18]. Epigenetic silencing of CDKN2A (p16) has been observed in early pre-neoplastic lesions [19, 20]. Based on the above literature, it is clear that the genetic features of GBC change with respect to the stage of cancer and this difference may explain the poor prognosis in advanced cases.
Any mutation in the KRAS oncogene that causes the altered expression of translated protein can cause cancer formation. In the literature, KRAS mutations have been reported in leukaemia, colorectal cancer, [10] pancreatic cancer [21], and lung cancer [22]. A previous study by Roa J C et al has found KRAS codon 12 mutations in 30% of GBC cases, which is consistent with our results [23]. However, Ajiki T et al., reported a higher incidence of KRAS mutations (i.e. 59%) [24]. The published literature shows that the frequency of KRAS mutations in GBC ranges from 8% to 80% which indicates the heterogenic nature of cancer [23–29].
The PIK3CA gene has also been found to be oncogenic and is well implicated in many cancers, like cervical cancer [30], breast cancer [31]. In the present study, we have sequenced exon 20 of the PIK3CA gene. The incidence of somatic pathogenic mutations present in PIK3CA exon 20 was 20% (14.7% have H1047R and 5.8% have H1047L change). A study performed in the Chilean population detected PIK3CA exon 20 (both H1047R and H1047L) mutations in 37.4% of GBC cases [32]. A research article published by Deshpande et al found PIK3CA exon 9 mutations in 18.8% GBC [33]. Another study by Zhao S et al reported the lower incidence of 6.15 % of PIK3CA exon 9 mutation in GBC patients [34]. However, in the last two articles, authors have not found any PIK3CA exon 20 mutations in GBC cases.
Similarly, previous studies have shown that mutations in the NRAS gene, which lead to a constantly active NRAS protein, are found in approximately ~13–25% of metastatic melanoma patients [35–37]. In the present study, we found NRAS exon 1 mutations in 3/34 (8.8%) of GBC cases. A previous study by Deshpande et al found NRAS exon 1 mutations in 6.3% of GBC cases [33].
Mutations that lead to EGFR overexpression (known as upregulation) or overactivity have also been reported previously in various cancers including GBC with variable frequencies like squamous-cell carcinoma of the lung (80% of cases), glioblastoma (50%) and epithelial tumours of the head and neck (80–100%) [38]. EGFR overexpression has also been reported in 87.5% GBC patients of North American ethnicity [39]. The somatic mutations involving EGFR can lead to its constant activation causing abnormal expression of EGFR protein which ultimately drives the neoplastic conversion of a cell. [15]. Our study has found EGFR exon 20 mutation in 7/34 (20.6%) cases.
Mutation in IDH2 has been frequently found in cancers like gliomas and acute myeloid leukemia. In the present study, we found IDH2 gene mutation in 4/34 (11.7%) of cases. The literature has not found much support for its association with GBC till now. Milind Javle et al has found IDH 1 gene mutation in 4/57 (7%) of GBC cases [40]. But on the contrary, a study by Darrell R. Borger et al found IDH gene hotspot mutations in cholangiocarcinoma but not in GBC [41].
The plausible explanation behind the variable results reported by various previous studies may be due to the Inter and intra-tumoural heterogeneity of cancer, which describes the observation of different tumour cells showing distinct morphological and molecular profiles including variable gene expression.
The role of mutations in cancer is well defined in the literature. Mutations are also designated as a hallmark of cancer, which allows a cell to invade and metastasise to other body parts. Multiple subtypes of a histopathologically common cancer make it crucial to understand the commonalities and differences among various types and subtypes. Presently, we had studied the distributions of mutation frequencies in five major cancer-related genes and establish their links to treatment outcome. In light of this, our study illustrates that despite being a histologically common entity of GBC (adenocarcinoma of gallbladder), there is heterogeneity with respect to the presence of mutations. KRAS, PIK3CA, and EGFR gene mutations were more common in GBC as compared to NRAS and IDH2. Moreover, KRAS mutations were also found to be significantly associated with an advanced stage of GBC.
The present study is based on a mutational analysis of five major cancer-related genes. We used micro-dissected gallbladder tissue with greater than 80% of tumour cells as a study material that imparts a positive strength to our results; still we had the limitation of a small sample size. Future studies with a higher number of samples are needed to better define genetically distinct subsets of cancers.
Conclusion
The present study found the presence of somatic mutations in KRAS, NRAS, PIK3CA, IDH2, and EGFR in GBC. None of these mutations were significantly associated with clinicopathological parameter except for KRAS exon 1 and 2 mutations with advanced stage of GBC. A more robust and larger study is further required to validate these findings.
Conflict of interest
The authors declare no conflict of interest.
Author contribution
Aarti Sharma and Dr Ashok Kumar designed, carried out the study and wrote the manuscript. Dr Niraj Kumari and Dr Narendra Krishnani provided the histopathological specimens and helped in tumour area micro-dissection work. Dr Neeraj Rastogi provided the clinical data.
Acknowledgments
The authors are very thankful to Dr Shubha Phadke, Professor and Head, Department of Medical Genetics, SGPGIMS, Lucknow, India, for providing the laboratory facilities.
References
1. Lazcano-Ponce EC, Miquel JF and Munoz N, et al (2001) Epidemiology and molecular pathology of gallbladder cancer CA Cancer J Clin 51(6) 349–364 https://doi.org/10.3322/canjclin.51.6.349
2. Randi G, Franceschi S, La Vecchia C (2006) Gallbladder cancer worldwide: geographical distribution and risk factors Int J Cancer 118(7) 1591–1602 https://doi.org/10.1002/ijc.21683 PMID: 16397865
3. Pandey M (2003) Risk factors for gallbladder cancer: a reappraisal Eur J Cancer Prev 12(1) 15–24 https://doi.org/10.1097/00008469-200302000-00004 PMID: 12548106
4. Henson DE, Albores-Saavedra J, Corle D (1992) Carcinoma of the gallbladder Histologic types, stage of disease, grade, and survival rates Cancer 70(6) 1493–1497 PMID: 1516000
5. Lai CH, Lau WY (2008) Gallbladder cancer–a comprehensive review Surgeon 6(2) 101–110 https://doi.org/10.1016/S1479-666X(08)80073-X PMID: 18488776
6. Pao W, Miller V, Zakowski M, et al (2004) EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib Proc Natl Acad Sci U S A 101(36) 13306–13311 https://doi.org/10.1073/pnas.0405220101 PMID: 15329413 PMCID: 516528
7. Paez JG, Janne PA and Lee JC, et al (2004) EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy Science 304(5676) 1497–1500 https://doi.org/10.1126/science.1099314 PMID: 15118125
8. Inoue A, Yoshida K and Morita S, et al (2016) Characteristics and overall survival of EGFR mutation-positive non-small cell lung cancer treated with EGFR tyrosine kinase inhibitors: a retrospective analysis for 1660 Japanese patients Jpn J Clin Oncol 46(5) 462–467 https://doi.org/10.1093/jjco/hyw014 PMID: 26977054 PMCID: 4874470
9. Sequist LV, Joshi VA and Janne PA, et al (2007) Response to treatment and survival of patients with non-small cell lung cancer undergoing somatic EGFR mutation testing Oncologist 12(1) 90–98 https://doi.org/10.1634/theoncologist.12-1-90 PMID: 17285735
10. Burmer GC, Loeb LA (1989) Mutations in the KRAS2 oncogene during progressive stages of human colon carcinoma Proc Natl Acad Sci U S A 86(7) 2403–2407 https://doi.org/10.1073/pnas.86.7.2403 PMID: 2648401 PMCID: 286921
11. Chang SC, Lin PC and Lin JK, et al (2016) Mutation spectra of common cancer-associated genes in different phenotypes of colorectal carcinoma without distant metastasis Ann Surg Oncol 23(3) 849–855 https://doi.org/10.1245/s10434-015-4899-z
12. Jang EK, Kim WG and Kim EY, et al (2016) Usefulness of NRAS codon 61 mutation analysis and core needle biopsy for the diagnosis of thyroid nodules previously diagnosed as atypia of undetermined significance Endocrine 52(2) 305–312 https://doi.org/10.1007/s12020-015-0773-9
13. Salvesen HB, Werner HM and Krakstad C, (2013) PI3K pathway in gynecologic malignancies Am Soc Clin Oncol Educ Book https://doi.org/10.1200/EdBook_AM.2013.33.e218 PMID: 23714506
14. Molenaar RJ, Radivoyevitch T and Maciejewski JP, et al (2014) The driver and passenger effects of isocitrate dehydrogenase 1 and 2 mutations in oncogenesis and survival prolongation Biochim Biophys Acta 1846(2) 326–341 PMID: 24880135
15. Lynch TJ, Bell DW and Sordella R, et al (2004) Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib N Engl J Med 350(21) 2129–2139 https://doi.org/10.1056/NEJMoa040938 PMID: 15118073
16. Barreto SG, Dutt A Chaudhary A (2014) A genetic model for gallbladder carcinogenesis and its dissemination Ann Oncol 25(6) 1086–1097 https://doi.org/10.1093/annonc/mdu006 PMID: 24705974 PMCID: 4037856
17. Wistuba II, Gazdar AF (2004) Gallbladder cancer: lessons from a rare tumour Nat Rev Cancer 4(9) 695–706 https://doi.org/10.1038/nrc1429 PMID: 15343276
18. Moreno M, Pimentel F and Gazdar AF, et al (2005) TP53 abnormalities are frequent and early events in the sequential pathogenesis of gallbladder carcinoma Ann Hepatol 4(3) 192–199 PMID: 16177659
19. Espinoza JA, Bizama C and García P, et al (2016) The inflammatory inception of gallbladder cancer Biochim Biophys Acta 1865(2) 245–254 PMID: 26980625
20. Tadokoro H, Shigihara T and Ikeda T, et al (2007) Two distinct pathways of p16 gene inactivation in gallbladder cancer World J Gastroenterol 13(47) 6396–6403 https://doi.org/10.3748/wjg.v13.i47.6396 PMID: 18081229 PMCID: 4205459
21. Almoguera C, Shiobata D and Forrester K, et al (1988) Most human carcinomas of the exocrine pancreas contain mutant c-K-ras genes Cell 53(4) 549–554 https://doi.org/10.1016/0092-8674(88)90571-5 PMID: 2453289
22. Tam IY, Chung LP and Suen WS, et al (2006) Distinct epidermal growth factor receptor and KRAS mutation patterns in non-small cell lung cancer patients with different tobacco exposure and clinicopathologic features Clin Cancer Res 12(5) 1647–1653 https://doi.org/10.1158/1078-0432.CCR-05-1981 PMID: 16533793
23. Roa JC, Roa I and de Aretxabata X, et al (2004) [K-ras gene mutation in gallbladder carcinoma] Rev Med Chil 132(8) 955–960 PMID: 15478297
24. Ajiki T, Fujimori T and Onoyama H, et al (1996) K-ras gene mutation in gall bladder carcinomas and dysplasia Gut 38(3) 426–429 PMID: 8675098 PMCID: 1383074
25. Singh MK, Chetri K and Pandey UB, et al (2004) Mutational spectrum of K-ras oncogene among Indian patients with gallbladder cancer J Gastroenterol Hepatol 19(8) 916–921 https://doi.org/10.1111/j.1440-1746.2004.03355.x PMID: 15242496
26. Takada M, Horita Y and Okuda S, et al (2002) Genetic analysis of xanthogranulomatous cholecystitis: precancerous lesion of gallbladder cancer? Hepatogastroenterology 49(46) 935–937 PMID: 12143246
27. Roa JC, Roa I and de Aretxabata X, et al (2005) Frequency of K-ras mutation in biliary and pancreatic tumors Rev Med Chil 133(12) 1434–1440
28. Tada M, Yokosuka O and Omata M, et al (1990) Analysis of ras gene mutations in biliary and pancreatic tumors by polymerase chain reaction and direct sequencing Cancer 66(5) 930–935 PMID: 2167148
29. Hanada K, Tsuchida A and Iwao T, et al (1999) Gene mutations of K-ras in gallbladder mucosae and gallbladder carcinoma with an anomalous junction of the pancreaticobiliary duct Am J Gastroenterol 94(6) 1638–1642 https://doi.org/10.1111/j.1572-0241.1999.01155.x PMID: 10364037
30. Ma YY, Wei SJ and Lin YC, et al (2000) PIK3CA as an oncogene in cervical cancer Oncogene 19(23) 2739–2744 https://doi.org/10.1038/sj.onc.1203597 PMID: 10851074
31. Karakas B, Bachman KE and Park BH (2006) Mutation of the PIK3CA oncogene in human cancers Br J Cancer 94(4) 455–459 https://doi.org/10.1038/sj.bjc.6602970 PMID: 16449998 PMCID: 2361173
32. Roa I, Gorica H and Game A, et al (2016) Somatic mutations of PI3K in early and advanced gallbladder cancer: additional options for an orphan cancer J Mol Diagn 18(3) 388–394 https://doi.org/10.1016/j.jmoldx.2015.12.003 PMID: 26947513
33. Deshpande V, Nduaguba A and Zimmerman SM, et al (2011) Mutational profiling reveals PIK3CA mutations in gallbladder carcinoma BMC Cancer 11 60 https://doi.org/10.1186/1471-2407-11-60 PMID: 21303542 PMCID: 3045359
34. Zhao S, Cao Y and Liu SB, et al (2016) The E545K mutation of PIK3CA promotes gallbladder carcinoma progression through enhanced binding to EGFR J Exp Clin Cancer Res 35(1) 97 https://doi.org/10.1186/s13046-016-0370-7 PMID: 27317099 PMCID: 4912708
35. Ball NJ, Yohn JJ and Morelli JG, et al (1994) Ras mutations in human melanoma: a marker of malignant progression J Invest Dermatol 102(3) 285–290 https://doi.org/10.1111/1523-1747.ep12371783 PMID: 8120410
36. Curtin JA, Fridlyand J and Kageshita T et al (2005) Distinct sets of genetic alterations in melanoma N Engl J Med 353(20) 2135–2147 https://doi.org/10.1056/NEJMoa050092 PMID: 16291983
37. van ‘t Veer LJ, Burgering BM and Versteeg R, et al (1989) N-ras mutations in human cutaneous melanoma from sun-exposed body sites Mol Cell Biol 9(7) 3114–3116 https://doi.org/10.1128/MCB.9.7.3114 PMID: 2674680 PMCID: 362784
38. Kumar V, Abbas A, Aster J (2013) Robbins basic pathology Philadelphia: Elsevier/Saunders p. 179 ISBN 9781437717815
39. Kaufman M, Mehrotra B and Limaye S et al (2008) EGFR expression in gallbladder carcinoma in North America Int J Med Sci 5(5) 285–291 https://doi.org/10.7150/ijms.5.285 PMID: 18825277 PMCID: 2556051
40. Javle M, Rashid A and Churi C et al (2014) Molecular characterization of gallbladder cancer using somatic mutation profiling Hum Pathol 45(4) 701–708 https://doi.org/10.1016/j.humpath.2013.11.001 PMID: 24508317 PMCID: 4428571
41. Borger DR, Tanabe KK and Fan KC, et al (2012) Frequent mutation of isocitrate dehydrogenase (IDH)1 and IDH2 in cholangiocarcinoma identified through broad-based tumor genotyping Oncologist 17(1) 72–79 https://doi.org/10.1634/theoncologist.2011-0386 PMCID: 3267826