Next-generation sequencing in NSCLC and melanoma patients: a cost and budget impact analysis
Rosa A van Amerongen1, Valesca P Retèl2,3, Veerle MH Coupé1, Petra M Nederlof4, Maartje J Vogel4 and Wim H van Harten2,3
1Department of Epidemiology and Biostatistics, VU University Medical Center, PO Box 7057, 1007 MB Amsterdam, The Netherlands
2Department of Psychosocial Research and Epidemiology, The Netherlands Cancer Institute, PO Box 90203, 1006 BE Amsterdam, The Netherlands
3School of Governance and Management, University of Twente, MB-HTSR, PO Box 217, 7500 AE Enschede, The Netherlands
4Department of Molecular Diagnostics, Pathology, The Netherlands Cancer Institute, PO Box 90203, 1006 BE Amsterdam, The Netherlands
Correspondence to: Valesca P. Retèl. Email: v.retel@nki.nl
Abstract
Next-generation sequencing (NGS) has reached the molecular diagnostic laboratories. Although the NGS technology aims to improve the effectiveness of therapies by selecting the most promising therapy, concerns are that NGS testing is expensive and that the ‘benefits’ are not yet in relation to these costs. In this study, we give an estimation of the costs and an institutional and national budget impact of various types of NGS tests in non-small-cell lung cancer (NSCLC) and melanoma patients within The Netherlands. First, an activity-based costing (ABC) analysis has been conducted on the costs of two examples of NGS panels (small- and medium-targeted gene panel (TGP)) based on data of The Netherlands Cancer Institute (NKI). Second, we performed a budget impact analysis (BIA) to estimate the current (2015) and future (2020) budget impact of NGS on molecular diagnostics for NSCLC and melanoma patients in The Netherlands. Literature, expert opinions, and a data set of patients within the NKI (n = 172) have been included in the BIA. Based on our analysis, we expect that the NGS test cost concerns will be limited. In the current situation, NGS can indeed result in higher diagnostic test costs, which is mainly related to required additional tests besides the small TGP. However, in the future, we expect that the use of whole-genome sequencing (WGS) will increase, for which it is expected that additional tests can be (partly) avoided. Although the current clinical benefits are expected to be limited, the research potentials of NGS are already an important advantage.
Keywords: Next-generation sequencing, test costs, budget impact, melanoma, NSCLC, personalised medicine, targeted therapy
Copyright: © the authors; licensee ecancermedicalscience. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/3.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Published: 28/10/2016; Received: 24/05/2016
Background
Next-generation sequencing (NGS) has reached the molecular diagnostic laboratories and is expected to (slowly) replace single-gene molecular diagnostic tests for the detection of DNA mutations in tumour tissues [1]. Within The Netherlands, currently, many (specialised) hospitals and university medical centres have implemented NGS for diagnostics in the clinic. NGS can be used to identify multiple specific mutations in a tumour, to which genomic-directed targeted therapy (hereinafter referred to as targeted therapy) may be assigned, as standard-of-care or referral to a clinical trial. A NGS panel can be described as ‘a multiplex predictive test, which explores limited regions of tumour DNA/RNA for aberrations that can be used as a molecular target for therapy’ [2]. There are several types, such as single-gene tests, small- (~50 genes, using most often a polymerase chain reaction (PCR)-based technique), medium- (~200 genes, using DNA captures), and large (using e.g., whole-exome sequencing (WES))-targeted gene panels (TGP), and whole-genome sequencing (WGS) [3]. The tests vary in price and turnaround time, and the design of the panels determines the different targeted therapy options.
There are high expectations around NGS and targeted therapies, they can improve patient survival and can reduce unnecessary harms and costs by preventing sequential single-gene testing and ineffective treatments [4–5]. For research, NGS is expected to speed up patient selection in clinical trials and to increase knowledge concerning the prognostic impact of targets [6–7]. In The Netherlands, an initiative was started by the Hartwig Medical Foundation (HMF) within the ‘Centre for Personalised Cancer Treatment (CPCT)’. The CPCT has the mission to serve all Dutch cancer patients with testing WGS for diagnostics. The CPCT is a nationally operating consortium involving all nine academic medical centres in The Netherlands (all UMC’s and The Netherlands Cancer Institute (NKI)) and an increasing number of teaching hospitals (STZ). The ultimate goal is to serve both direct patient diagnostics towards experimental care (clinical trials, off-label drug use) and the construction of a nationwide database with sequencing and treatment response information.
Currently, molecular diagnostics and targeted therapies are mostly recommended for patients in an advanced stage of disease. Especially, for stage-IV non-small-cell lung cancer (NSCLC) and stage-IV melanoma patients, NGS can be very beneficial due to their heterogeneous nature, high mutation frequency, and the availability of targeted therapies. For both patient groups, the prognosis is poor, stage-IV NSCLC patients have a 5-year survival rate of 1% and stage-IV melanoma patients of 15–20% [8–9]. Since 2004 for NSCLC and 2011 for melanoma, several targeted therapies are approved by the European Medicines Agency (EMA) [10]. These therapies show promising results, overall response rates above 60% are measured in both NSCLC and melanoma patients treated with targeted therapy, compared with 20% by chemotherapy [11–12]. In case of no actionable mutation, which is currently the case in approximately 78% of the NSCLC and 49% of the melanoma patients, usual chemotherapy or immunotherapy is recommended [13]. In the current Dutch diagnostic guidelines, still only the testing of two single genes are recommended for stage-IV NSCLC patients (epidermal growth factor receptor (EGFR) and the anaplastic lymphoma kinase (ALK)) [14]. For stage-IV melanoma patients, so far, no molecular diagnostic test is recommended, although a single-gene test investigating the BRAF V600E-gene (BRAF) mutation status is often applied [15]. However, as seen in the NKI, specialised hospitals already use more extensive molecular diagnostic tests for both patient groups.
Although the NGS technology aims to improve the effectiveness of therapies, concerns are that NGS testing is expensive and that the clinical benefits are not yet in relation to these costs [16]. Currently, there are publications on cost-effectiveness analyses of NGS available [17–18]. However, these publications do not contain detailed cost analyses regarding the sequencing tests themselves. In order to support the decision whether the implementation of NGS should be prolonged or extended within the clinic, our department started an early health technology assessment (HTA), with a scenario analysis focused on NGS implementation by means of expert elicitation [2]. Subsequently, explorations of the potential costs and benefits are needed, starting with the costs of NGS and expected developments. The objective of the current study was firstly, to calculate the costs for the small- and medium TGP used within our institute (NKI) and secondly, to estimate the budget impact of the old, current and future situation around NGS and other molecular diagnostic test costs from a national perspective (Dutch), and an institutional perspective (NKI).
Methods
Activity-based costing: test costs
The total costs of performing two types of NGS tests for diagnostics have been analysed for a small TGP (48 genes) and a medium TGP (178 genes). For this, we used activity-based costing (ABC), a cost calculation technique that links resource costs with products using a multistep allocation procedure on the basis of activity consumption [19]. This practically means that for each product, in this case a diagnostic test, all required activities that consume resources, varying from consumables to personnel time, are included in the calculation. The cost calculations for the two types of TGPs were based on NKI data. A small TGP, targeting 48 genes and 212 amplicons on DNA level (the TruSeq Amplicon Cancer Panel (TSACP) from Illumina BV), was recently implemented in routine diagnostics. The medium TGP was an in-house developed capture panel covering the full exons of 178 cancer-related genes, using a customised SureSelect XT2 capture library and magnetic streptavidin beads for capture. This panel can be used for the detection of both DNA and RNA variants, but in this study, only the DNA part is considered. Both the procedures can be executed on the Illumina MiseqTM and the Illumina HiseqTM, and per run a maximum of 48 samples can be included, except for a run with the medium TGP on the MiseqTM which is restricted to four samples per run. These maximums of four or 48 samples per run are related to the desired minimum depth of coverage and pipetting time needed per sample.
The test costs consisted of the following elements: personnel, material, equipment, and an estimation of the overhead costs. Personnel costs were based on the estimated minutes spent per process step, both per sample and run. Hour rates per personnel type were based on the NKI employers’ costs of 2014 and only the direct productive hours were included in this calculation. Real ‘timing activity’ was used to calculate the time required for the small TGP currently in clinical practice. Since the medium TGP was not used in a diagnostic setting, timing estimations were made by means of interviews with the executers of the process. Material costs were based on laboratory protocols and purchase prices within the NKI. Laboratory protocols were also used to calculate the equipment costs; realistic redemption durations and yearly service costs were taken into account. All costs were divided into fixed yearly costs, fixed costs per run and variable costs per sample. The cost consequences of important parameters of the NGS procedure were explicitly investigated; the type of sequence technology (Illumina MiseqTM/HiseqTM), the numbers of samples per run, the number of runs per week and the completeness of the run. Variation in these parameters will result in different molecular diagnostic test costs and turnaround times of the diagnostic process.
Budget impact analysis
A budget impact analysis (BIA) was performed to estimate the yearly budget impact of NGS tests in NSCLC and melanoma patients from a Dutch perspective and an institutional perspective (NKI). A BIA focuses on the expected changes in the expenditure of a health care system after the adoption of a new intervention over a time period of 5 years in this case. Time frames 2012, 2015, and 2020 have been selected to show an estimation of the old, current and future budget impact. In 2012, mostly (a combination of) single tests and methods were used in clinical diagnostics. In 2015, a variety of NGS tests were represented from single-gene tests in the general hospitals, till small-medium TGPs in the academic centres. In the future time frame of 2020, it was assumed that WGS will be the clinical standard in academic/specialised hospitals by that time. The latter assumption is in the framework of the CPCT, their mission is to perform WGS in all specialised centres, for both diagnostics and research.
Within our BIA, six sources of input are included: three test cost sources, literature for diagnostic guidelines and patient numbers, patient data for the case study and expert opinions. The BIA has been performed according to the BIA framework of the International Society for Pharmacoeconomics and Outcomes Research (ISPOR) [20]. This framework consists of several standard aspects: target population, scenario distribution based on hospital types, resource utilisation, costs per unit, total costs, and sensitivity analyses. Our BIA model is shown in Figure 1 and the different aspects are discussed below.
Target population and scenario distribution (nationwide)
The NSCLC and melanoma incidence within The Netherlands and the stage distribution at diagnosis were used to calculate the size of the target population, stage IV NSCLC and melanoma patients within The Netherlands. The patients were divided into two groups: peripheral or specialised healthcare (the latter including NKI, university medical centres and other specialised hospitals), according to the number of patients diagnosed per hospital type. Different molecular diagnostic test standards have been taken into account for these two types of hospitals. We assumed that for each new stage-IV patient, molecular diagnostics is executed once. Although repeated testing over time is often required in case of progression, among others caused by tumour heterogeneity within a patient, we assumed no influence by NGS on this frequency.
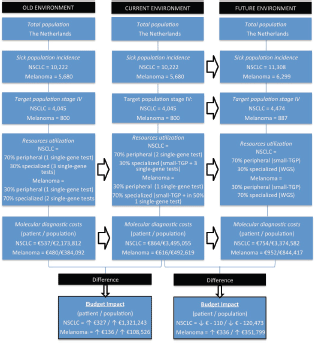
Figure 1. Budget impact analysis model, showing the aspects within the ISPOR framework for the old, current, and future environment. The arrows indicate a change of one of the aspects overtime. The target populations are divided into patients receiving peripheral and specialized care, based on the type of hospital they are diagnosed. At the bottom, the average cost differences per patient and population between the old and current environment and current and future environment are shown for both NSCLC and melanoma patients. NSCLC = non-small-cell lung cancer, NGS = next-generation sequencing, WGS = whole-genome sequencing.
Resource utilisation
The resource utilisation within this BIA reflects the use of molecular diagnostic tests (including personnel, material, and equipment use) within the Dutch health care system. Based on the Dutch diagnostic guidelines [14–15], the clinical standard of molecular diagnostics in peripheral hospitals has been estimated. The two used diagnostic guidelines for NSCLC patients for the old and current situation were implemented in, respectively, 2011 and 2015. For melanoma patients, no molecular diagnostic test is included in the guideline yet; however, since 2012 a single-gene test is recommended, and therefore, we assume this test to be the clinical standard in peripheral hospitals. We made the assumption that in all specialised hospitals the old and current clinical standard of molecular diagnostics was similar as within the NKI. The test scenarios in the future situation, including expected patient numbers for which NGS is requested, process improvements and renewed gene panels, are based on expectations within the NKI. We assume in our analysis that no other changes in the general executed diagnostics and imaging will occur. We also assume biopsy- and test failure rates to stay similar in the future.
Costs per unit
Three different sources have been used to estimate the molecular diagnostic test costs. (1) The costs for single-gene tests were estimated within the NKI and it was assumed that these costs are comparable in other hospitals. (2) The small- and medium-TGP costs were based on the ABC analysis explained in the “Activity-based costing: test costs” section. (3) The WGS costs estimation was based on the public data of the National Human Genome Research Institute, showing the most recent sequence costs per DNA sequence or genome [21], and the future estimation was discussed with experts in The Netherlands. We assumed that the current costs of molecular diagnostic tests in clinical practice are representative for future costs as well. However, for the WGS costs, currently still under development with major cost reductions over the past years, we assume a reduction of 75% in five years.
Sensitivity analysis
Considering the fact that many included parameters for the BIA were based on hypothetical values and extrapolations, plausible ranges for the model’s parameters were explored. We conducted two one-way sensitivity analyses, one for the test costs in the current NGS situation of NSCLC and one for the influence of future test costs on the budget impact. We varied key parameters one-by-one, using several uncertainty ranges. For the test costs, four parameters have been included: (1) Number of NGS samples per week, (2) Number of NGS runs per week, (3) Completeness of the NGS run, and (4) Required additional tests besides NGS. All base values and ranges were based on realistic possibilities within the NKI. Three treatment parameters were included in the sensitivity analysis for the future budget impact: (1) Price of TGP using NGS, (2) Price of WGS, and (3) Percentage of patients that will receive specialised care (using WGS testing). For these parameters, a range of 20% has been used, as seen in other comparable BIA’s [22].
Institutional perspective (case study): BIA within the NKI
In order to analyse the budget impact of NGS in clinical practice, the NKI was used as a case study environment. Stage-IV NSCLC and melanoma patients for who molecular diagnostics were requested in a period before NGS implementation (November–December 2014) and in a period after NGS implementation (January–February 2015) were included. A data set was created with the diagnosis, number, and type of molecular diagnostic tests per patient and the observed mutations per test. Both actionable and non-actionable mutations were counted and the percentages of patients with an actionable mutation for which hypothetically a targeted therapy is available (EMA approved or in trial) were calculated. Treatment consequences were manually checked via the electronic health records of the patients and four categories were added to the data set: (1) whether there was a treatment option for the patient, (2) which treatment was selected, (3) whether the selected treatment was based on the test result, and (4) whether the patient received a first dose of the treatment. The molecular diagnostic tests, test results, and treatment consequences have been compared for the two periods. Based on the molecular tests executed per patient, the test costs per patient and for the total NKI population were calculated, including a budget impact of the NGS implementation. We assumed that the number of included patients in these 3.5 months was representative for the rest of the year, in order to estimate the yearly NKI population of NSCLC and melanoma patients for who molecular diagnostic tests are requested.
Analysis
Microsoft Excel and SPSS 23.0 have been used to perform the sensitivity analyses and to create descriptive statistics of the results. An independent two-sample t-test for the continuous variables with a normal distribution, an independent samples Mann–Whitney U-test for the continuous variables without a normal distribution and a Fisher’s exact test for the binary variables have been used to analyse differences between the subgroups of the NKI case study. P values below 0.05 were considered to indicate a statistically significant difference.
Results
Activity-based costing: NGS costs
Figure 2 shows the estimated costs per sample dependent on the number of samples per run. A higher number of samples per run resulted in lower costs per sample, due to the fixed costs per year and fixed costs per run. The following mean costs per sample are given for a run with 24 samples, one run executed per week and on average 85% of the run filled. Between brackets the minimum costs with 48 samples and maximum costs with four samples are shown, respectively. Costs per sample with the small TGP were €606 (€465; €1769) on the MiseqTM and €956 (€621; €4,161) on the HiseqTM. A run with the medium TGP on the MiseqTM is restricted to four samples per run, for €3009 per sample, and costs per sample on the HiseqTM were €1,137 (€857; €5273). These costs are inclusive VAT, overhead (30%) and the costs of control samples, in a run with ≤8 samples one control sample is included and in a run with ≥9 samples two control samples are included. A general overview of the costs is shown in Table 1 and Table S1 and S2 give a more comprehensive overview of the included costs.
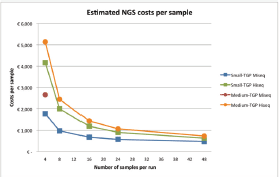
Figure 2. Estimated costs per sample, dependent on the number of samples per run, assuming one run per week, and complete runs. The costs are shown for both the small- and medium TGP on the Illumina MiseqTM or HiseqTM. There is a maximum of four samples for the medium TGP on the Illumina MiseqTM, therefore only one data point is given. NGS = next-generation sequencing, TGP = targeted gene panel.
Table 1. A summary of the NGS costs for four options; small or medium TGP on the Illumina MiseqTM or HiseqTM.
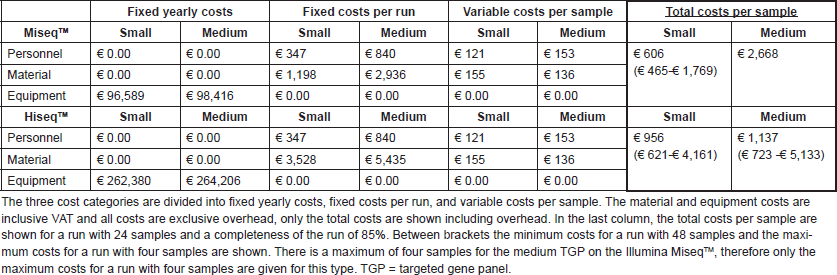
Table S1. Overview of the NGS test costs with the small TGP.
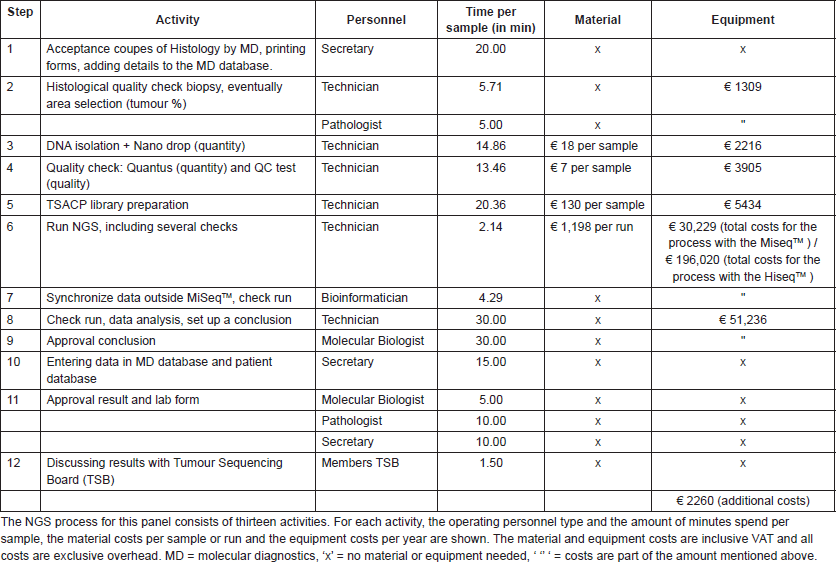
Table S2. Overview of the NGS test costs with the medium TGP. The NGS process for this panel consists of fourteen activities.
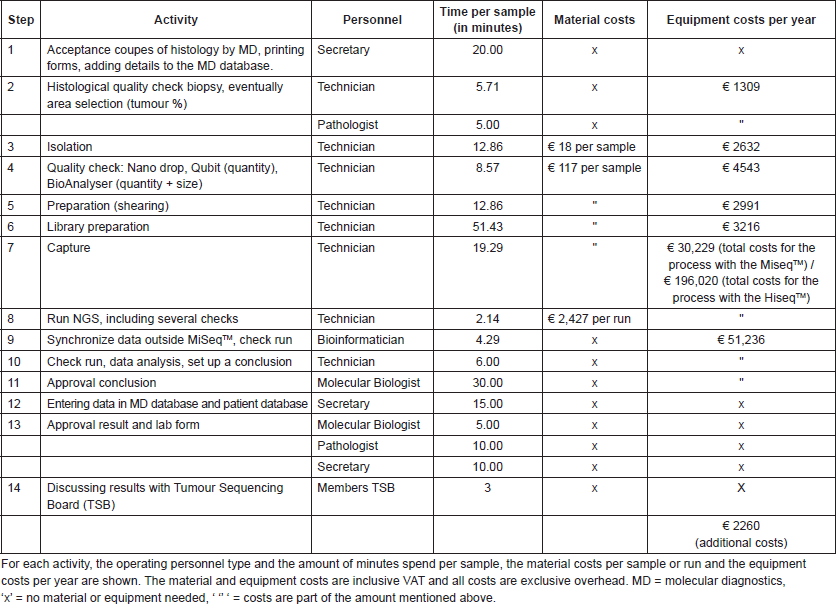
Budget impact analysis
Target population and scenario distribution (nationwide)
Within The Netherlands, the current incidence of stage-IV NSCLC and melanoma patients are, respectively, 4,045 and 800 patients per year [23–25]. In the future situation of our BIA, 4,474 NSCLC and 887 melanoma patients are included, based on the expected increase of 11% in 5 years [26]. We assumed that 70% of the NSCLC patients receive their care in a general hospital, the other 30% of patients receive specialised care [24]. Since 2013 it has been determined that all stage-IV melanoma patients have to be referred to specialised melanoma centres (14 specialised hospitals in The Netherlands). This results in a distribution where 70% of the patients are treated in a specialised hospital and 30% in a peripheral hospital [27].
Resource utilisation
An overview of the specific test types per time frame and hospital type, including the costs per unit as discussed in the next part, is given in Table 2. In the old situation, in peripheral hospitals, we assumed one single-gene test as clinical standard for both NSCLC (EGFR) and melanoma patients (BRAF). In the current situation for NSCLC, we assumed one additional single-gene test (ALK), based on the current guidelines [14–15]. In the old situation within specialised hospitals, we assumed that multiple single-gene tests were performed, minimally three for NSCLC patients (a multigene panel, HER2/EGFR, and ALK, ROS, RET, or MET) and two for melanoma patients (BRAF and NRAS or KIT). In the current situation in specialised hospitals, small–medium TGPs are increasingly implemented. Based on our experience in the NKI, it is estimated that, especially for NSCLC, additional tests will be required due to (temporary) inefficiencies and due to specific translocations not included in the small TGP. Therefore, besides the small TGP, three additional single-gene tests were assumed for NSCLC patients (e.g., fragment analysis (HER2/EGFR) to detect larger deletions/insertions, Sanger sequencing (EGFR) and fluorescence in situ hybridisation (ALK, ROS, RET, or MET)) and one single-gene test was assumed for 50% of the melanoma patients (Sanger Sequencing (KIT)). In the future (2020), we assumed in our model that NGS with a small TGP will be clinical practice in peripheral hospitals for both NSCLC and melanoma patients. In specialised hospitals, we assumed WGS to be clinical practice, following the developments of the CPCT. For both NGS and WGS, we assumed in the future no required additional tests caused by inefficiencies.
Costs per unit
In peripheral hospitals, the average diagnostic test costs per NSCLC patient in the old, current, and future situation were, respectively, €295; €590, and €606. In the specialised hospitals, the test costs were, respectively, €1103; €1504, and €1100. For melanoma patients in peripheral hospitals, these costs were, respectively, €295, €295, and €606 and in specialised hospitals €559, €753, and €1100. In Table S3, an overview of the costs per test is shown, including the cost source and an explanation for the WGS costs estimation.
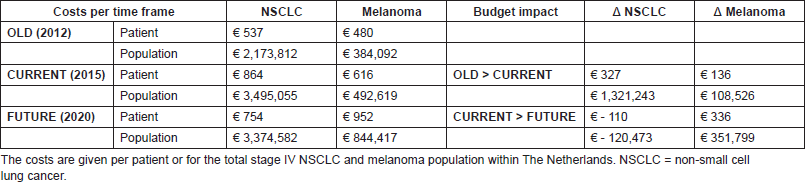
Total costs
The yearly budget impact for NSCLC patients was as follows: the current situation will result in a test cost increase of €327 per patient, followed by a cost decrease of €110 in the future situation. For the total population in The Netherlands, this will be, respectively, an annual increase in €1,321,243 and annual decrease in €120,473. For melanoma patients, an increase of €136 and €336, respectively, in test costs per patient is expected, resulting in a yearly budget impact of €108,526 and €351,799 for the total population. A summary of the BIA results is given in Table 3.
Table 2. The molecular diagnostic tests for NSCLC and melanoma patients are shown per hospital type in the old, current and future situation.
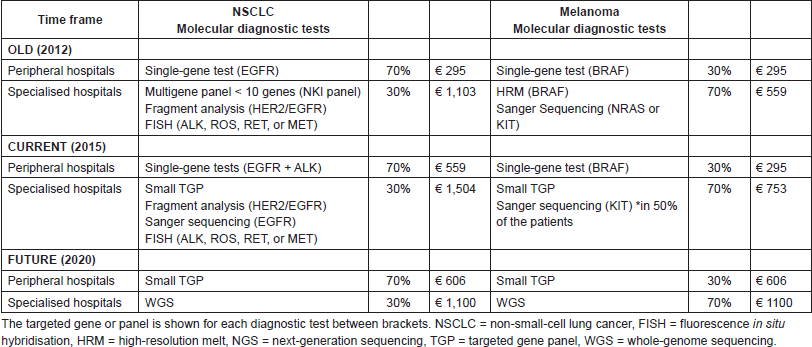
Table S3. Overview of the molecular diagnostic test costs within the BIA.
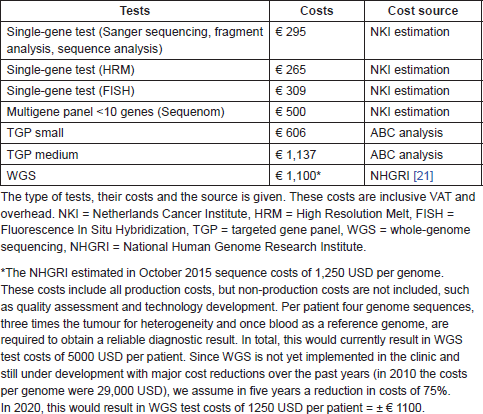
Table 3. Final BIA results, showing the test costs per time frame and showing the budget impact of the current and future situation.
Sensitivity analyses
The sensitivity analysis of the test costs in the current NGS situation of NSLCC in specialised hospitals showed a range of €606 to €2614. For the base value, €1504 as shown in the ‘costs per unit’ section, it is assumed that per NGS run 24 samples are included, per week one run is executed, on average 85% of the run is filled and three additional single gene tests are required. The biggest reduction in test costs can be achieved by a reduction in additional tests besides NGS. The base value, range, and the effect on the test costs are shown for each parameter in the tornado plot in Figure 3. The sensitivity analysis of the budget impact of the future scenario compared with the current situation shows for the NSCLC population a range of minus €500,080 to plus €259,134. For the melanoma population a range between €215,173 and €488,424 has been calculated. The base values, minus €120,473 (NSCLC) and €351,799 (melanoma), are based on the results of our BIA calculation, discussed in the ‘total costs’ section. The price of NGS shows the widest range in the budget impact for the NSCLC population. For the melanoma population, the biggest reduction can be achieved by a reduction in the price of WGS. The tornado plots of these analyses are shown in Figure 4.
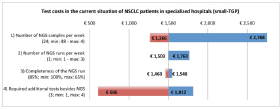
Figure 3. One-way sensitivity analysis on the effect of four parameters on the test costs in the current situation of NSCLC patients in specialised hospitals. TGP = targeted gene panel.
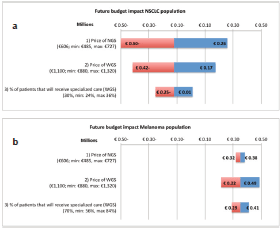
Figure 4a and b. One-way sensitivity analysis on the effect of three parameters on the budget impact of the future scenario. The amounts on the x-as are shown in millions and account for the Dutch NSCLC patient population (4a) and the Dutch melanoma patient population (4b). NSCLC = non-small-cell lung cancer, NGS = next-generation sequencing, WGS = whole-genome sequencing.
Institutional perspective (case study): BIA within the NKI
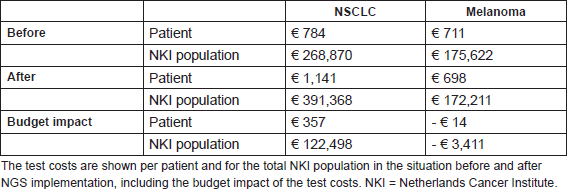
In total 172 patients were included, all stage-IV NSCLC and melanoma patients within the chosen time periods have been selected. The test-related measures, mutation, and treatment characteristics for the period before and after NGS implementation are shown in Table 4. In NSCLC patients, there is only a significant increase in the number of tests and tests costs after the implementation of NGS. In melanoma patients, significant differences have been observed both in test-related measures and mutation characteristics: the number of tests per patient dropped after the NGS implementation and the number of observed mutations increased. The test costs per patient and for the total patient population in the period before and after NGS implementation are shown in Table 5. Yearly 343 stage-IV NSCLC patients and 247 stage-IV melanoma patients within the NKI were assumed. The NGS implementation resulted within the NKI in a budget impact of an additional spending of €357 per NSCLC patient and a saving of €14 per melanoma patient. For the total NKI population, this meant a budget impact of €119,087 (€122,498 increase for NSCLC patients and €3,411 savings for melanoma patients).
Table 4. Test-related measures, mutation, and treatment characteristics for NSCLC and melanoma patients within the NKI, for whom molecular diagnostic tests were executed in 2 months before NGS implementation and 1.5 months after NGS implementation.
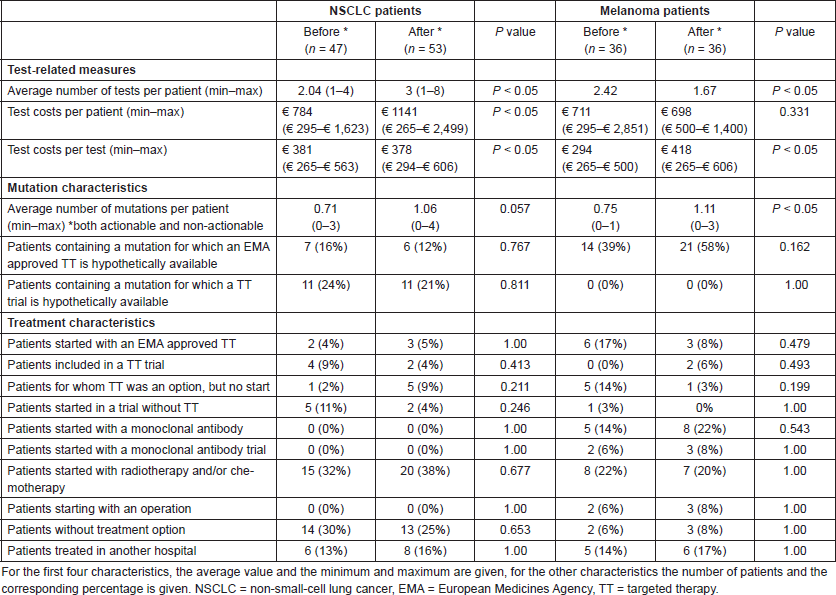
Table 5. Summary of the BIA case study.
Discussion
As far as we know, this is one of the first published cost analysis and budget impact analysis regarding NGS. As expected, our results showed that the more samples are included and runs are performed, the less costly NGS will become. Regarding budget impact, we showed that currently NGS resulted in an increase in test costs, from 2012 to 2015 yearly €1.32 million for the total NSCLC population and €0.11 million for the total melanoma population. The finding of these current budget impacts was confirmed by the NKI case study. From the current to future situation, our BIA showed for the total NSCLC population an expected annual decrease of €0.12 and for the total melanoma population an increase of €0.35 million. This decrease in NSCLC patients was related to the more efficient use of testing by WGS in specialised hospitals, without the need of additional tests. The increase in future test costs for melanoma patients was related to both the implementation of NGS in peripheral hospitals and the implementation of WGS in specialised hospitals, while no additional test costs were involved. Our sensitivity analyses produced widely varying budget impact estimates in the future; especially the price of the small TGP in NSCLC and the price of WGS in melanoma patients had a high impact.
The ABC analysis produced a wide range for the estimate of NGS test costs depending on the number of samples per run, number of runs per week, device, and type of TGP. These calculated test costs are comparable with other recently published cost analysis of NGS techniques. For example, in a situation with six samples per run 600–900 USD was calculated for a similar small TGP and 2000 USD for a medium TGP [28]. Policy decisions in institutions have to be made which panel type and number of runs per week will be most beneficial for patients, regarding both the costs and consequences. For example, the decision whether an increase in the number of mutations tested can outweigh the higher costs and turnaround time. In a macro level, although the current costs in combination with the benefits of NGS are still unclear, NGS especially has a lot of potential for the future. Firstly, NGS is expected to replace single-gene tests, which can reduce the costs and limits patient material use [5]. Currently, additional tests are still needed to supplement the small TGP test; however, it is expected that eventually these additional tests become (partly) superfluous. The research potentials of NGS are an important potential advantage as well [6–7]. A broader panel of genes can result in more treatment options for patients, both off-label and in clinical trial. This can speed up patient recruitment for clinical trials and consequently targeted therapies can become EMA approved more rapidly. These potentials are relevant both for current and future patients.
Our analyses have some limitations. Firstly, we made several assumptions in the BIA. In the future situation, we assumed no inefficiencies of NGS or WGS that require additional tests and no findings of new clinical relevant modifications not traceable with WGS. We also assumed that the current costs of molecular diagnostic tests in clinical practice are representative for the future and we have made an estimation for the WGS costs reduction in five years. The second limitation of our BIA is the fact that the consequences of NGS are not taken into account; in particular, treatments costs can have a major impact on the total budget. By identifying more targetable targets, NGS can result in an increased use of targeted therapies [29]. However, without clinical data, it is very complicated to estimate the treatment consequences. In clinical practice, patients receive several treatment lines and several factors, besides mutation status, play a role in therapy selection or whether a treatment is prescribed at all [30]. Furthermore, it is hard to estimate the availability of new targeted therapies, their prices and average treatment durations in the future. The third limitation of our study is related to the generalisability of our NKI case study and the clinical practice within the NKI, which we used to estimate the test situation in specialisd hospitals within The Netherlands. The NKI is a specialised comprehensive cancer centre, with a broader variety of molecular diagnostic tests and a higher number of clinical trials. The patient population is different within the NKI as well; patients are often redirected to the NKI after an ineffective first-line treatment elsewhere. In addition, a larger data set would have been desirable, however, this was not feasible due to the recent implementation.
Nevertheless, the NKI case study showed interesting and comparable results with our main Dutch BIA. Between the period before and after NGS implementation, only differences in average number of tests and associated costs were found. No significant differences in mutation and treatment characteristics were found, except for the number of observed mutations in melanoma patients. We expected beforehand that no difference in EMA approved targeted therapy use would be observed, since the number of druggable targets is low and within the time period no additional targeted therapies were EMA approved [10]. However, an increase in participation in trials would have been a realistic option, since off label-targeted therapy use is investigated in several trials. Not finding an increase so far can hypothetically be caused by the small data set in combination with the strict inclusion criteria for trials and the recent implementation of NGS, whereby trials are not yet focused on new NGS results. Although no direct clinical benefits of NGS have been measured, the increase in number of observed mutations in melanoma (significant) and NSCLC patients (not significant) is already in line with the expected research potentials of NGS. Furthermore, the test costs in the NKI case study and the main Dutch BIA in specialised hospitals are comparable: per NSCLC patient, respectively, €784 versus €1103 in the old situation and €1141 versus €1504 in the current situation. This applies for the melanoma patients as well, per patient, respectively, €711 versus €559 in the current situation and €698 versus €753 in the future situation. An explanation for the difference in test costs in the current situation is related to the fact that not for all patients in the NKI case study NGS was executed. The selected molecular diagnostic tests are dependent on the request of the clinician or on already known mutation profiles of patients due to previously executed molecular tests.
Conclusions and implications
Based on our analysis, we expect that the concerns, of NGS resulting in high additional test costs, will be limited. Our BIA does show that NGS currently results in higher costs, which is mainly related to the required additional tests besides the small TGP. However, it is expected that these additional tests become (partly) superfluous in the future. Furthermore, the direct clinical benefits of NGS are currently limited, since no treatment consequences are yet expected due to the low number of druggable targets. Nonetheless, the research potentials of NGS are already an important advantage.
Our cost analysis of NGS and the NKI case study can assist hospitals in the decision which panel or device is most beneficial for them. Based on our analysis, we would suggest a small TGP as currently the best NGS option for diagnostics and we recommend NGS process optimisation to focus on the reduction of additional tests. Our research provides useful input for a health technology assessment of NGS. Interesting follow-up steps would be an extension of our BIA approach to the total cancer population within The Netherlands and the inclusion of clinical data including treatment costs. Furthermore, the inclusion of additional future scenarios with liquid biopsies or a centralised NGS test option within The Netherlands, as proposed by the CPCT/MHF, would be of interest as well.
List of abbreviations
NGS Next-generation sequencing
WGS Whole-genome sequencing
WES Whole-exome sequencing
NSCLC Non-small-cell lung cancer
ABC Activity-based costing
BIA Budget impact analysis
TGP Targeted gene panel
NKI Netherlands Cancer Institute
DNA Deoxyribonucleid acid
CPCT Centre for Personalised Cancer Treatment
HMF Hartwig Medical Foundation
STZ Dutch teaching hospitals
UMC University Medical Centre
EMA European Medicines Agency
EGFR Epidermal growth factor receptor
ALK Anaplastic lymphoma kinase
BRAF Rapid accelerating fibrosarcoma type B oncogene B1
HER2 Human epidermal growth factor receptor 2
KRAS Kirsten-rat sarcoma 2 viral oncogene homolog
ROS receptor tyrosine kinase
RET rearranged during transfection
MET MNNG-HOS transforming gene
HTA Health technology assessment
TSACP TruSeq Amplicon Cancer Panel
ISPOR International Society for Pharmacoeconomics and Outcomes Research
Conflicts of Interest
Authors declare no conflict of interest regarding the work described in this manuscript.
Authors’ contributions
RVA held in-house interviews, collected the input for the cost analysis and BIA, processed the data, analysed the results, and drafted the manuscript. VR held in-house interviews, coordinated the input collection, analysed the results, and co-drafted the manuscript. VC helped to set up the BIA, advised on the results analysis, and critically revised the manuscript. PN provided information on the test activities, process costs, and patient data, gave insight in future expectations and critically revised the manuscript. MJ provided information on the test activities, gave insight in future expectations, and critically revised the manuscript. WVH conceived the work, coordinated the project and helped drafting the manuscript. All authors read, revised, and approved the manuscript.
Acknowledgments
We acknowledge the ‘NGS-project’ team from the NKI for their valuable input.
References
1. Vrijenhoek T, Kraaijeveld K and Elferink M et al (2015) Next-generation sequencing-based genome diagnostics across clinical genetics centers: implementation choices and their effects Eur J Hum Genet 23(9) 1142–1150 DOI: 10.1038/ejhg.2014.279 PMID: 25626705 PMCID: 4538197
2. Joosten SE, Retèl VP and Coupé VM et al (2016) Scenario drafting for early technology assessment of next generation sequencing in clinical oncology BMC Cancer 16(1) 66 DOI: 10.1186/s12885-016-2100-0 PMID: 26851938 PMCID: 4744630
3. Zhao X, Wang A and Walter V et al (2015) Combined targeted DNA sequencing in non-small cell lung cancer (NSCLC) using UNCseq and NGScopy, and RNA Sequencing Using UNCqeR for the detection of genetic aberrations in NSCLC PLoS One 10(6)
4. Diamandis M, White NM and Yousef GM (2010) Personalized medicine: marking a new epoch in cancer patient management Mol Cancer Res 8(9) 1175–1187 DOI: 10.1158/1541-7786.MCR-10-0264 PMID: 20693306
5. Hagemann IS, Devarakonda S and Lockwood CM et al (2015) Clinical next-generation sequencing in patients with non-small cell lung cancer Cancer 121(4) 631–639 DOI: 10.1002/cncr.29089
6. Gonzalez de castro D, Clarke PA and Al-lazikani B et al (2013) Personalized cancer medicine: molecular diagnostics, predictive biomarkers, and drug resistance Clin Pharmacol Ther 93 (3) 252–259 DOI: 10.1038/clpt.2012.237 PMID: 23361103 PMCID: 3577635
7. Frampton GM, Fichtenholtz A and Otto GA et al (2013) Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing Nat Biotechnol 31(11) 1023–1031 DOI: 10.1038/nbt.2696 PMID: 24142049
8. American Cancer Society Non-small cell lung cancer survival rates [available at : http://www.cancer.org/cancer/lungcancer-non-smallcell/detailedguide/non-small-cell-lung-cancer-survival-rates] Date accessed: 01-06-2015
9. American Cancer Society Melanoma skin cancer survival rates [available at: http://www.cancer.org/cancer/skincancer-melanoma/detailedguide/melanoma-skin-cancer-survival-rates] Date accessed: 01-06-2015
10. European Medicines Agency European public assessment reports [available at: http://www.ema.europa.eu/ema/] Date accessed: 01-06-2015
11. Kumarakulasinghe NB, Van zanwijk N and Soo RA (2015) Molecular targeted therapy in the treatment of advanced stage non-small cell lung cancer (NSCLC) Respirology 20 (3) 370–378 DOI: 10.1111/resp.12490 PMID: 25689095
12. Michielin O and Hoeller C (2015) Gaining momentum: New options and opportunities for the treatment of advanced melanoma Cancer Treat Rev 41(8) 660–670 DOI: 10.1016/j.ctrv.2015.05.012 PMID: 26096079
13. Memorial Sloan-Kettering Cancer Center cBioPortal for Cancer Genomics [available at: http://www.cbioportal.org] Date accessed: 01-06-2015
14. Netherlands Comprehensive Cancer Organisation (IKNL) Oncoline treatment guideline NSCLC version 2.1 [available at: http://www.oncoline.nl/niet-kleincellig-longcarcinoom] Date accessed: 01-06-2015
15. Netherlands Comprehensive Cancer Organisation (IKNL) Oncoline treatment guideline Melanoma version 2.0 [available at: http://www.oncoline.nl/melanoom] Date accessed: 01-06-2015
16. Christensen KD, Dukhovny D and Siebert U et al (2015) Assessing the costs and cost-effectiveness of genomic sequencing J Pers Med 5(4) 470–486 DOI: 10.3390/jpm5040470 PMID: 26690481 PMCID: 4695866
17. Doble B, John T and Thomas D et al (2016) Cost-effectiveness of precision medicine in the fourth-line treatment of metastatic lung adenocarcinoma: an early decision analytic model of multiplex targeted sequencing. Lung Cancer In Press, Corrected Proof DOI: 10.1016/j.lungcan.2016.05.024
18. Li Y, Bare LA and Bender RA et al (2015) Cost-effectiveness of sequencing 34 cancer-associated genes as an aid for treatment selection in patients with metastatic melanoma Mol Diagn Ther 19(3) 169–177 DOI: 10.1007/s40291-015-0140-9 PMID: 25926090 PMCID: 4469775
19. Lievens Y, Van den bogaert W and Kesteloot K (2003) Activity-based costing: a practical model for cost calculation in radiotherapy Int J Radiat Oncol Biol Phys 57(2) 522–535 DOI: 10.1016/S0360-3016(03)00579-0 PMID: 12957266
20. Sullivan SD, Mauskopf JA and Augustovski F et al (2014) Budget impact analysis-principles of good practice: report of the ISPOR 2012 budget impact analysis good practice II task force Value Health 17(1) 5–14. DOI: 10.1016/j.jval.2013.08.2291 PMID: 24438712
21. Wetterstrand KA DNA sequencing costs: data from the NHGRI genome sequencing program (GSP) [available at: www.genome.gov/sequencingcosts] Date accessed: 01-11-2015
22. Bajaj PS, Veenstra DL and Goertz HP et al (2014) Targeted erlotinib for first-line treatment of advanced non-small cell lung cancer: a budget impact analysis J Med Econ 17(8) 538–546 DOI: 10.3111/13696998.2014.912987 PMID: 24716717
23. Netherlands Comprehensive Cancer Organisation (IKNL) The Netherlands Cancer Registry [available at: http://www.cijfersoverkanker.nl] Date accessed: 01-06-2015
24. Signaleringscommissie Kanker van KWF Kankerbestrijding (2010) Kwaliteit van kankerzorg in Nederland, Oisterwijk.
25. Zorginstituut Nederland, Tumor infiltrerende lymfocyten bij patiënten met uitgezaaid melanoom irresectabel stadium IIIc en IV [available at: https://www.zorginstituutnederland.nl/pakket/lopende dossiers/voorwaardelijke toelating/tumor-infiltrerende-lymfocyten-bij-patienten-met-uitgezaaid-melanoom-irresectabel-stadium-iiic-en-iv.html] Date accessed: 01-06-2015
26. Signaleringscommissie Kanker van KWF Kankerbestrijding (2011) Kanker in Nederland tot 2020, Trends en prognoses Oisterwijk
27. Dutch Institute for Clinical Auditing (2015) Dutch melanoma treatment registry: Resultaten van behandeling met nieuwe geneesmiddelen bij gevorderd melanoom in Dutch Institute for Clinical Auditing (DICA) annual report 2014, Leiden
28. Sabatini LM, Mathews C and Ptak D et al (2016) Genomic sequencing procedure microcosting analysis and health economic cost-impact analysis: a report of the association for molecular pathology J Mol Diagn 18(3) 319–328 DOI: 10.1016/j.jmoldx.2015.11.010 PMID: 27080370
29. Kris MG, Johnson BE and Berry LD et al (2014) Using multiplexed assays of oncogenic drivers in lung cancers to select targeted drugs JAMA 311(19) 1998–2006 DOI: 10.1001/jama.2014.3741 PMID: 24846037 PMCID: 4163053
30. Frenkel M (2013) Refusing treatment Oncologist 18(5) 634–636 DOI: 10.1634/theoncologist.2012-0436 PMID: 23704223 PMCID: 3662856