Phosphate-to-alanine ratio and bilirubin-to-androsterone glucuronide ratio are the hub metabolites in upper gastrointestinal cancers: a Mendelian randomisation (MR) study
Pengkhun Nov1†, Duanyu Wang1†, Chongyang Zheng1, Syphanna Sou2, Socheat Touch2, Samnang Kouy2, Virak Vicheth1, Lilin Li1, Yangfeng Zhang1, Xiang Liu1, Changqian Wang1, Peizan Ni1, Qianzi Kou1, Ying Li1, Arzoo Prasai1, Wen Fu1, Wandan Li1, Kunpeng Du1,a and Jiqiang Li1,b
1Department of Radiation Oncology, Oncology Center, Zhujiang Hospital of Southern Medical University, Guangzhou, Guangdong Province 510282, China
2Department of Radiation Oncology and Oncology, Khmer-Soviet Friendship Hospital of University of Health Sciences, Phnom Penh 120110, Cambodia
ahttps://orcid.org/0000-0002-0684-7291
bhttps://orcid.org/0000-0002-585-5911
†These authors contributed equally to this work.
Abstract
Objective: Upper gastrointestinal (UGI) cancers, particularly esophageal cancer (EC) and gastric cancer (GC) represent a significant health burden with complex etiologies. Metabolic alterations are known to play a crucial role in cancer development and progression. Identifying key metabolic biomarkers may offer insights into the pathophysiology of UGI cancers and potential therapeutic targets. This study aimed to investigate the causal associations between 1,400 types of metabolites, specifically phosphate-to-alanine and bilirubin-to-androsterone glucuronide, and the risk of developing UGI cancers using Mendelian randomisation (MR) analysis.
Method: We conducted a two-sample MR study utilising genetic instruments identified from large-scale genome-wide association studies (GWASs) for metabolic traits. The outcomes were derived from GWAS datasets of UGI cancer patients, including EC and GC. Several MR methods were employed to ensure the robustness of the findings, including inverse variance weighted (IVW), MR-Egger and weighted median approaches.
Results: Our analysis found a total of 44 metabolites associated with EC and 15 metabolites associated with GC. The MR analyses revealed a significant causal relationship between the phosphate-to-alanine ratio (EC: OR = 1.002,95% CI = 1.00034−1.0037, p = 0.0037; GC: OR = 1.24,95% CI = 1.046−1.476, p = 0.01) and increased risk of UGI cancers. In contrast, the bilirubin-to-androsterone glucuronide ratio (EC: OR = 0.998,95% CI = 0.997−0.999, p = 0.03; GC: OR = 0.80,95% CI = 0.656−0.991, p = 0.04) was inversely associated with the risk, suggesting a potential protective effect.
Conclusion: Our findings suggest that the phosphate-to-alanine ratio and bilirubin-to-androsterone glucuronide ratio are key hub metabolites in the etiology of UGI cancers. These metabolic ratios could serve as potential biomarkers for early detection or targetsfor therapeutic intervention. Further research is warranted to elucidate the underlying biological mechanisms and to validate the clinical utility of these associations.
Keywords: esophageal cancer, gastric cancer, metabolites, mendelian randomisation (MR), genome wide association study (GWAS)
Correspondence to: Kunpeng Du and Jiqiang Li
Email: dkp321098@smu.edu.cn and ljq821028@126.com
Published: 23/07/2024
Received: 12/02/2024
Publication costs for this article were supported by ecancer (UK Charity number 1176307).
Copyright: © the authors; licensee ecancermedicalscience. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Introduction
Upper gastrointestinal (UGI) cancers, encompassing malignancies of the esophagus and stomach, remain a leading cause of cancer-related morbidity and mortality worldwide [1]. The prognosis in many countries remains dire, largely due to the absence of effective screening programs [2, 3]. Understanding the mechanisms underlying the initiation and progression of these cancers is crucial for developing effective prevention and treatment approaches. In recent years, there has been increasing evidence that metabolites, small molecules involved in cellular metabolism, are linked to the causal pathways of UGI cancers. Advances in molecular biology and the development of various omics approaches have significantly enhanced molecular epidemiological studies in this field [4].
An imbalance in metabolism is increasingly recognised as a crucial element in the development of UGI cancers [5]. Beyond the well-established alterations in glucose metabolism, such as the Warburg effect, disturbances in the metabolism of nucleotides, lipids and amino acids have also been reported in both lab and clinical research [6–8]. Metabolites, which are the end products of intricate interplays between inherent metabolic processes, genetic factors and environmental conditions, act as a snapshot of these biochemical processes. Metabolomics, utilising high-throughput techniques, allows for the detailed detection and quantification of a vast range of small-molecule metabolites (with molecular weights less than 1,000 Da) from a single sample. This approach is instrumental in discovering new biomarkers and in deepening our understanding of the mechanisms of cancer development [9, 10]. Moreover, it paves the way for uncovering novel preventative strategies and therapeutic targets [11].
Previous observational studies have indicated a link between metabolites and an increased likelihood of developing UGI cancer [4, 12–15]. However, observational studies are susceptible to biases and confounding factors, limiting their ability to establish causality. Mendelian randomisation (MR) was initially conceptualised and developed as a method to provide robust evidence of causality through the analysis of genetic variation in relation to exposure and outcome, serving as a potential alternative to randomised controlled trials [16]. This approach leverages the random allocation of genetic variation at conception, well before the onset of disease, making MR a valuable tool for establishing causality and mitigating the risk of reverse causality, independent of confounders typically present in study designs [17]. Here, we utilised MR to investigate the histophysiology and pathophysiological involvement of the metabolites in the development of digestive tract cancers, achieved by a recent statistical summary from a genome-wide association study (GWAS) focused on metabolites [18].
The phosphate-to-alanine ratio and the bilirubin-to-androsterone glucuronide ratio have been implicated in various metabolic processes relevant to cancer biology. Phosphate is essential for energy metabolism and cellular signaling, while alanine, a non-essential amino acid, can be utilised by cancer cells to support gluconeogenesis and energy production under hypoxic conditions [33, 41]. Bilirubin, a product of heme catabolism, exhibits antioxidant properties, whereas androsterone glucuronide, a metabolite of steroid hormones, may influence cancer growth through hormonal pathways [60, 63]. This MR study aims to systematically investigate the causal relationships between 1,400 types of metabolites, specifically phosphate-to-alanine and bilirubin-to-androsterone glucuronide, and the risk of developing UGI cancers. By doing so, we seek to elucidate the potential of these ratios as hub metabolites in the metabolic landscape of UGI cancers and to explore their utility as biomarkers for cancer risk or progression. The findings could provide a foundation for novel preventive and therapeutic strategies targeting metabolic dysregulation in UGI malignancies.
Materials and methods
Study design
The relationship between a vast array of metabolites and UGI cancers was examined in this study through the application of two-sample MR analyses. MR exploits genetic variants as surrogates for modifiable risk factors to infer causality. For MR to provide trustworthy conclusions about causal relationships, it is imperative that the instrumental variables (IVs) employed meet three critical criteria: First, the genetic variant must have a direct association with the metabolite of interest (the exposure). Second, the genetic variant should not be associated with confounding factors that could influence both the exposure and the outcome, ensuring that observed associations are not spurious. Finally, the genetic variant should affect the outcome solely through its impact on the exposure, with no alternative pathways influencing the outcome Figure 1.
Data sources for exposure and outcome
The sources of metabolite-wide GWAS data. The statistics summary of GWAS for each metabolite is publicly available from the European GWASs (accession number: GCST90199621-90201020) or non-European GWASs (accession number: GCST90201021-90204063) [19]. To identify relevant data for each cancer type, we used cancer-specific keywords to search (https://gwas.mrcieu.ac.uk/). For esophageal cancer (EC), we selected ieu-b-4960 (EC), and for gastric cancer (GC), we chose ebi-a-GCST90018849 (GC). Following this, we downloaded the corresponding data (from https://www.ebi.ac.uk/gwas/) based on the IDs of each cancer type, which was used for the analysis of the relation between 1,400 types of metabolites and UGI cancers. A reference panel derived from Sardinian sequences was utilised [20] to estimate approximately 22 million single-nucleotide polymorphisms (SNPs) genotyped using high-density arrays, and correlations were assessed following adjustment for covariates. The GWAS database is a comprehensive collection of genetic variation and its association with various traits or diseases. It provides a valuable resource for researchers and clinicians interested in understanding the genetic basis of complex traits and diseases. Based on the ID of each cancer, we used online data from GWAS including 372,756 European individuals (n = 740 case patients and 372,016 control participants) for EC, and 476,116 European individuals (n = 1,029 case patients and 475,087 control participants) for GC to analyse the relationship between 1,400 type metabolites and each cancer according to IDs (https://www.ebi.ac.uk/gwas/).
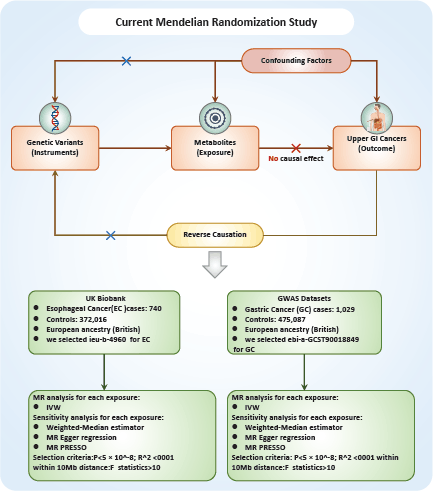
Figure 1. Flowchart of the study design.
Instrument selection
In light of the large number of SNPs reaching genome-wide significance (p < 5 × 10-8) for metabolite traits, we adopted more stringent criteria (p < 5 × 10-9) for selecting genetic IVs [21]. These IVs were pinpointed by categorising them based on the linkage disequilibrium reference panel from the 1,000 Genomes Project, applying a cutoff of R2 < 0.001 at a distance of up to 1,000 kilobases (kb). Due to the comparatively smaller size of GWAS datasets for metabolites, we utilised a p-value threshold of 5 × 10-8 and a more relaxed clustering threshold (R2 < 0.1 at a distance of 500 kb) [22]. To ensure the strength of our genetic instruments, we only selected IVs with F-statistics greater than 10, thereby qualifying them as robust instruments for our analyses. These IVs were then extracted from the summary data for UGI cancer outcomes. We excluded any SNPs that demonstrated potential pleiotropic effects (p < 10-5) on UGI cancer, which is consistent with protocols from prior studies [23]. We harmonised the SNPs across the datasets for exposures and outcomes to ensure coherent effect size estimations for the same alleles. SNPs with effect allele frequencies greater than 0.42 or those that were incompatible with harmonisation, were omitted from our analysis [22]. This process of careful SNP selection and harmonisation ensures the integrity and consistency of our MR analysis.
Statistical analysis
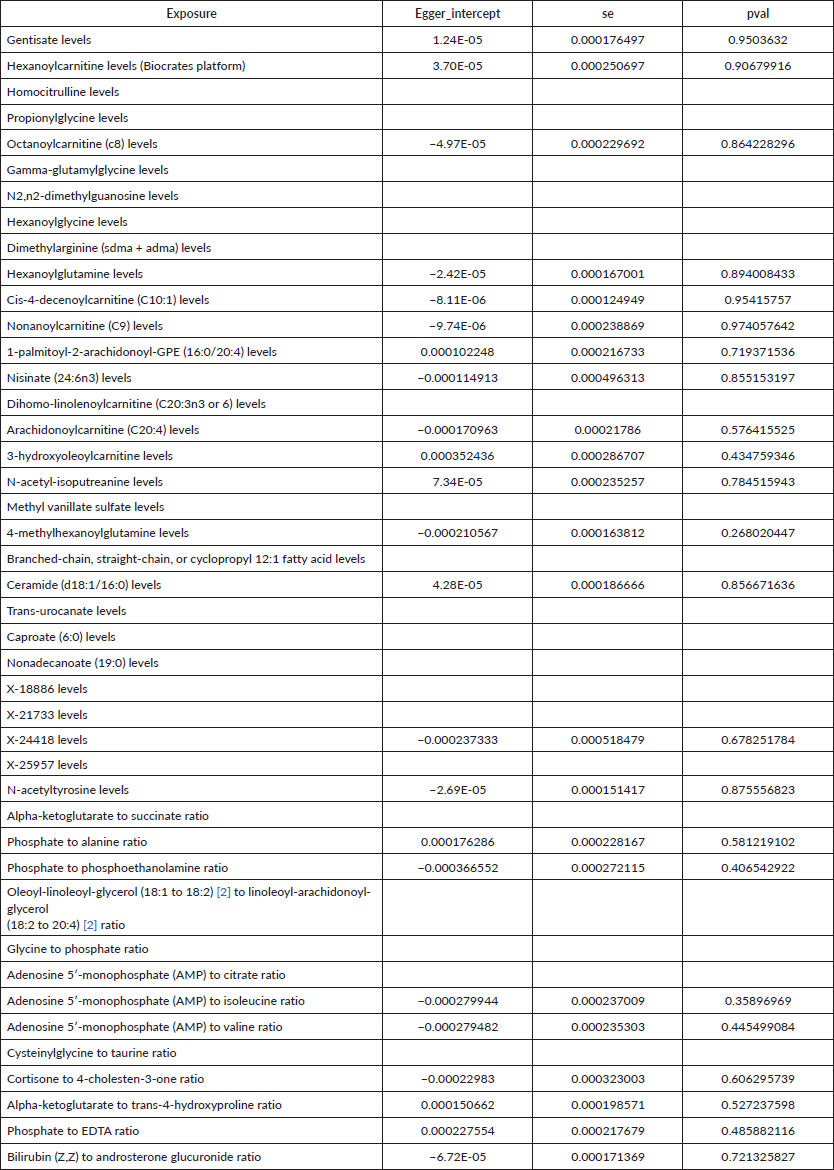
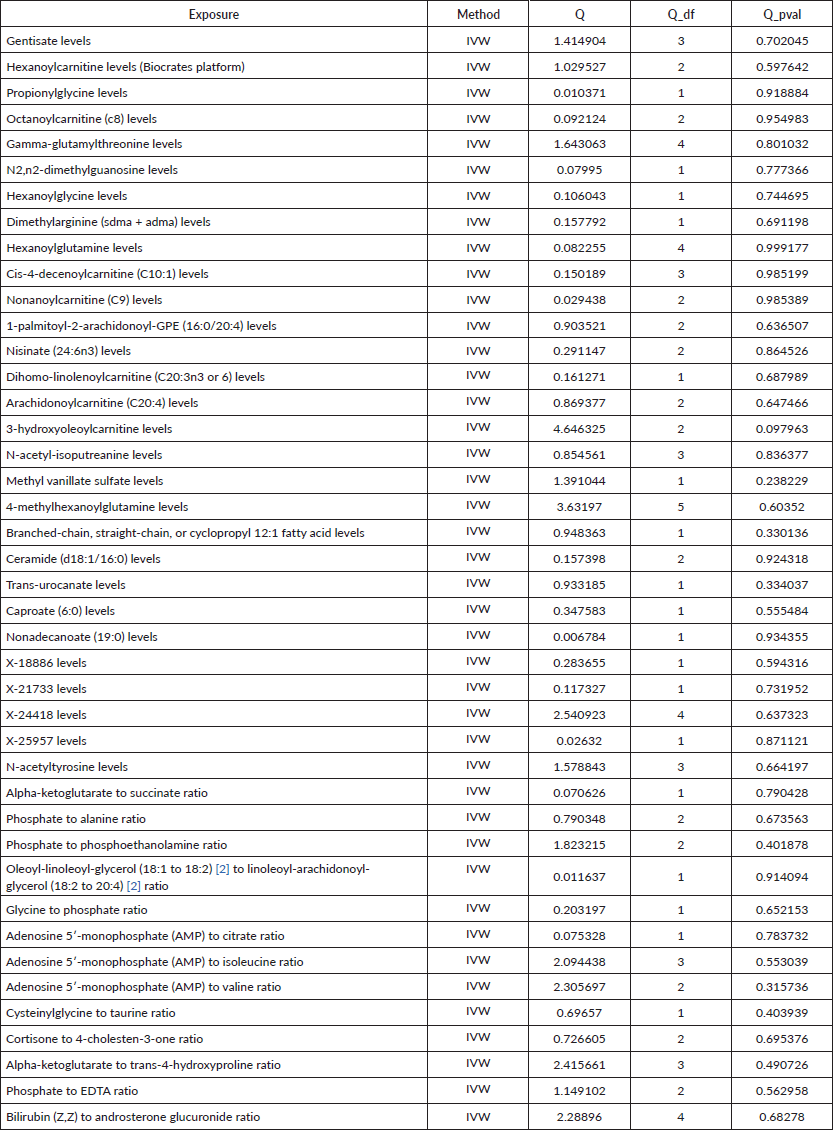
R 4.3.1 software (http://www.Rproject.org) was used to perform the analysis. Three methods were primarily utilised to reveal the causal relationship between 1,400 types of metabolites and UGI cancers: inverse variance weighting (IVW) [24], median-based weighting [25] and pattern-based weighting [26]. These analysis were primarily carried out using the software package of ‘TwoSampleMR’ (version 0.4.3) [27]. To assess heterogeneity among the selected IVs, Cochran's Q statistical along with corresponding values were utilised. Where the null hypothesis was rejected, we opted for random effects IVW in lieu of fixed effects [24]. In addressing the potential impact of horizontal pleiotropy, we employed the MR-Egger method. This method is particularly useful for detecting the presence of horizontal pleiotropy, indicated by a statistically significant intercept term [24]. Furthermore, we used the MR Pleiotropy Residual Sum and Outlier method, a robust approach to identify and eliminate potential horizontal pleiotropic outliers that might significantly influence our estimation results [28]. To further validate our findings, scatterplots and funnel plots were used. These plots confirmed that our results were not biased by outliers and demonstrated that the correlations observed were robust and exhibited no significant heterogeneity.
Results
To assess the causal impact of various metabolites on EC and GC, we employed a two-sample MR analysis using the method of IVW. In our study, we found a total of 44 metabolites associated with EC and 15 metabolites associated with GC (Tables 1 and 2). Phosphate-to-alanine ratio (EC: OR = 1.002,95% CI = 1.00034− 1.0037, p = 0.0037; GC: OR = 1.24,95% CI = 1.046− 1.476, p = 0.01) and bilirubin (Z,Z) to androsterone glucuronide (EC: OR = 0.998,95% CI = 0.997− 0.999, p = 0.03; GC: OR = 0.80,95% CI = 0.656− 0.991, p = 0.04) are common metabolites of EC and GC. Among them, phosphate-to-alanine ratio are risk factor for GC and EC, while bilirubin (Z,Z) to androsterone glucuronide are protective factors for GC and EC. Our findings are summarised in Figures 2 and 3. However, neither the MR-Egger intercept test nor Cochran's Q test revealed pleiotropy and heterogeneity (Supplementary Tables 1 and 2).
Discussion
MR has become a pivotal tool for demonstrating potential causal risk factors in diseases. In the present study, we used MR to establish an inverse causal relationship between metabolites and UGI cancers.
Table 1. Causal estimation between metabolites and EC.
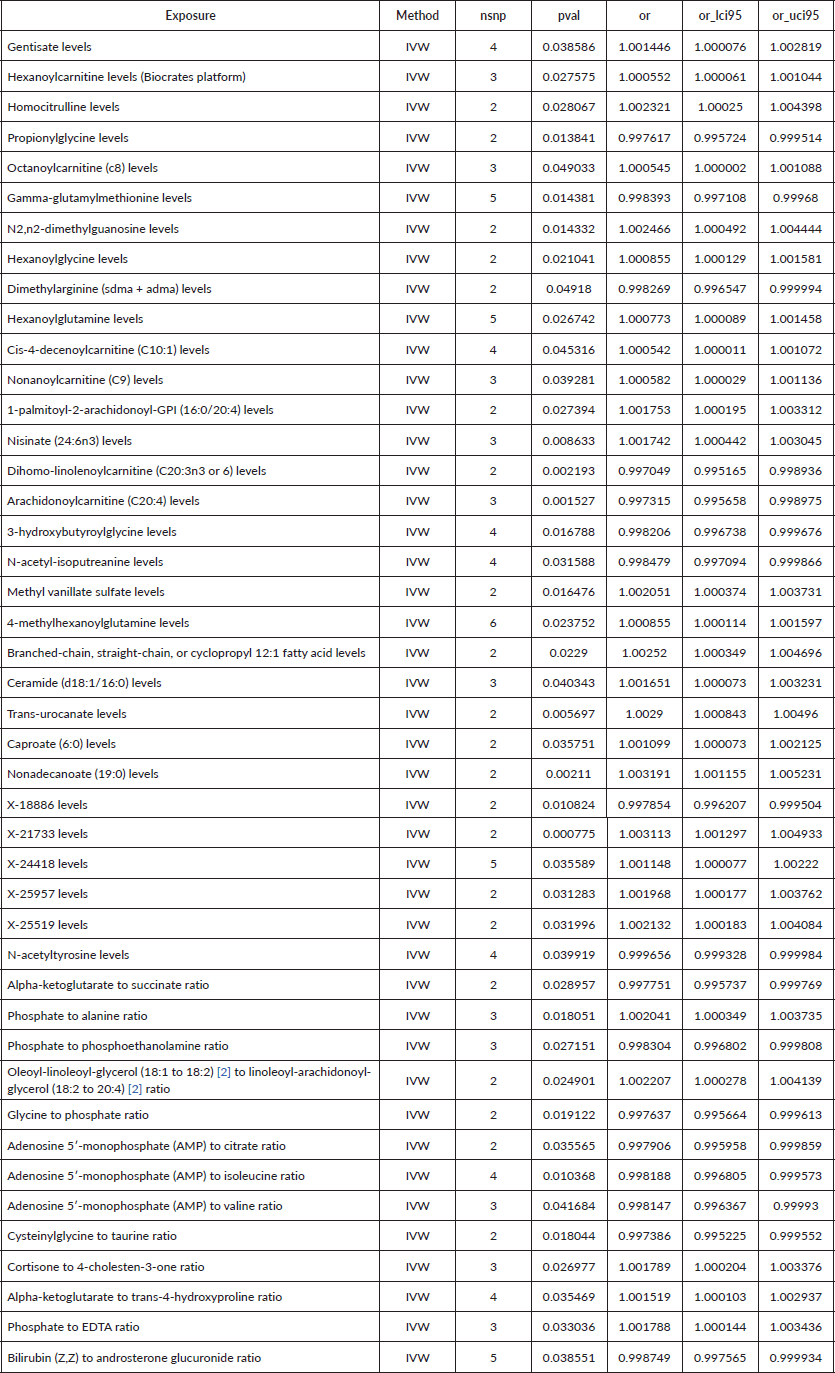
Table 2. Causal estimation between metabolites and GC.
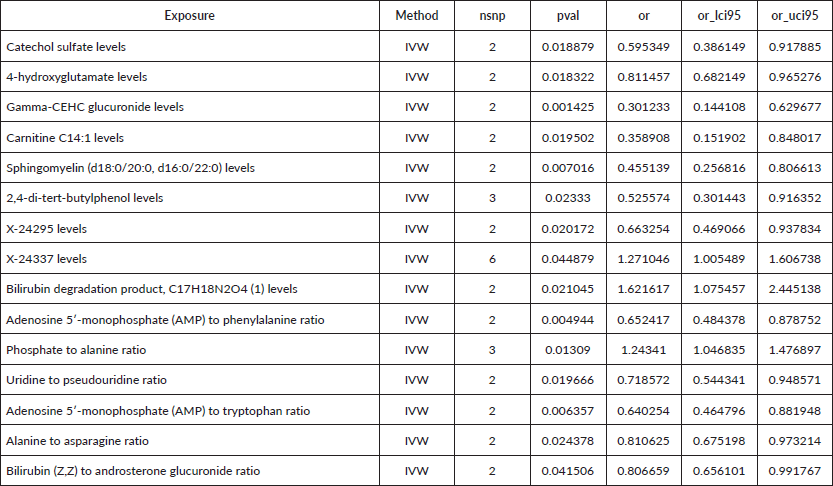

Figure 2. Causal estimation between metabolites and EC.

Figure 3. Causal estimation between metabolites and GC.
The nexus between metabolites and cancer has garnered significant attention recently. The development and progression of cancer are intrinsically linked to cellular metabolism. Cancer cells often exhibit unique metabolic pathways, such as relying on glycolysis and lactic acid fermentation for glucose metabolism instead of mitochondrial pathways [29]. Additionally, tumours may reprogram lipid metabolism, enhancing lipid uptake and accumulation, which alters the tumour microenvironment, suppresses immune responses and promotes tumour progression [30]. The complexity of tumour metabolism and the heterogeneous role of metabolites in cancer make this a challenging area of study. Metabolites are not only predictors of cancer risk but also play crucial roles in cancer treatment, where reprogramming metabolic pathways can help overcome chemotherapy resistance [31, 32]. In our MR analysis, we investigated the associations between serum metabolites and the risks of EC and GC. Our findings reveal distinct metabolite profiles associated with these cancers in the European population, as well as some common metabolites. This has led to the identification of novel candidate metabolites that may influence the risk of EC and GC. These insights offer new avenues for the treatment and management of these cancers, contributing significantly to our understanding of their etiology and mechanisms. In our investigation into EC cancer, we identified 44 metabolites related to the disease, while in GC cancer, we identified 15 related metabolites. Among them, phosphate-to-alanine ratio and bilirubin (Z, Z) to androsterone glucuronide ratio were found to be jointly connected to the risk of UGI cancers. Our findings suggest that a high phosphate-to-alanine ratio is linked to an increased risk of UGI cancers, whereas a higher bilirubin (Z, Z) to androsterone glucuronide ratio might act as a protective factor. Phosphate plays an important role in cell growth. For example, liver regeneration requires a large amount of phosphate to synthesize nucleotide triphosphate due to DNA replication. After hepatectomy, a decrease in phosphate indicates liver cell regeneration and a lack or delay of this decrease suggests impaired regeneration [33]. In small-cell lung cancer, diminished serum phosphate has been associated with an abnormal increase in fibroblast growth factor 23 [34]. Moreover, phosphates like nicotinamide adenine dinucleotide are central to maintaining redox homeostasis in cancer [35]. For instance, in gliomas with functional phosphate and tensin homologs, hypoxia induces endogenous production of cytoplasmic reactive oxygen species (ROS) and tumour cell growth by activating nicotinamide adenine dinucleotide phosphate (NADPH) oxidase, contributing to radiation resistance [36]. Lung cancer cells typically exhibit elevated ROS and NADPH levels [37], and NADPH is also associated with pancreatic cancer risk [38]. Research by Shi et al [39] indicated that Myoferlin disrupts the redox balance, promotes ROS production and increases the ratio of NADPH/NADP+, thereby accelerating the metastasis of GC. These studies corroborate our findings that elevated phosphate levels, possibly through increased NADPH production, lead to enhanced ROS production, promoting tumour cell proliferation and metastasis in UGI cancers. A critical aspect of cancer cell interaction with their environment involves the exchange of metabolites, especially amino acids [40]. A significant portion of metabolic alterations in cancer relates to amino acid metabolism and biosynthesis. Alanine is a non-essential amino acid primarily synthesized in the mitochondrial matrix. Alanine aminotransferase competes with pyruvate dehydrogenase (PDH), and PDH oxidizes decarboxylation to form acetyl CoA. PDH is responsible for the oxidative decarboxylation of pyruvate to form acetyl CoA, a process that is subdued under hypoxic conditions, leading to increased alanine synthesis [41]. In hepatocellular carcinoma, the alanine metabolism pathway undergoes significant changes typically marked by a reduction in alanine content [42]. Interestingly, in breast cancer, β- Alanine and alanine have the same chemical formula but differ in structure. The increase in their content due to a decrease of 4-aminobutyrate aminotransferase normally catalyzes β- Alanine into malonic acid semialdehyde, which can be converted into acetyl coenzyme A by simultaneously reducing NAD+to NADH [43]. Furthermore, the synthesis of alanine in breast cancer is influenced by the α-ketoglutarate demand, driven by pyruvate uptake in the tumour microenvironment [44]. In our study, we also observed a correlation between the high risk of UGI cancers and reduced alanine levels, which is consistent with the findings in hepatocellular carcinoma. This suggests that the metabolic pathway of alanine in gastrointestinal tumours might follow a similar pattern, with cancer cells potentially absorbing and deaminating alanine to form pyruvate, an essential carbon source for synthesizing other compounds. Thus, our results indicate that the phosphate-to-alanine ratio is linked to the increased risk of UGI cancers. The interaction between phosphate and alanine and the impact of their relative concentrations could serve as a potential predictive biomarker for UGI cancers. Bilirubin, the end product of red blood cell degradation, is categorised into direct bilirubin (DBIL) and indirect bilirubin (IBIL). Previous studies have highlighted the antioxidant, anti-inflammatory and anticancer properties of bilirubin, especially IBIL [45–47]. In addition, studies have confirmed that high bilirubin is associated with favourable prognosis and lower incidence rates in other cancers, including lung, breast and colorectal cancer [48–50]. However, the study by Wei et al [51] found reduced levels of total bilirubin (TBIL), DBIL and IBIL in GC patients, leaving the causal relationship between bilirubin reduction and GC unclear. In esophageal squamous cell carcinoma, research by Huang et al [52] suggested that elevated preoperative serum bilirubin levels (including non-conjugated bilirubin, conjugated bilirubin and TBIL) have been linked to a longer overall OS. Typically, elevated serum bilirubin is an indicator of liver dysfunction [53]. Given the liver’s involvement in multiple metabolic processes, liver cancer patients often experience abnormalities in multiple liver function indicators in the late stage. Therefore, composite index ratios like the albumin bilirubin score (ALBI) have been increasingly used to evaluate liver cancer prognosis, demonstrating substantial predictive value [54]. ALBI has also been recognised as a prognostic factor for GC, EC, colon cancer, pancreatic cancer and non-small cell lung cancer [55–58]. A retrospective study involving 628 patients undergoing radical gastrectomy for GC showed that those with higher preoperative ALBI levels experienced a significantly higher incidence of postoperative complications [59]. Similarly, in patients with ampullary adenocarcinoma undergoing radical pancreaticoduodenectomy, a high preoperative TBIL to albumin ratio was identified as an independent protective factor against recurrence [60]. In EC, a higher incidence of postoperative anastomotic leakage was observed to be 46.3% in the high ALBI group versus 27.5% in the low ALBI group (p = 0.038) [58].
Steroid hormone metabolism typically activates pathways involved in cell proliferation, survival, migration and invasiveness and is related to cancer initiation and progression closely [61]. Androsterone glucuronide, a liver metabolite of a glucosyltransferase-modified steroid oestrogen and testosterone, as well as a metabolite of the steroid dihydrotestosterone found in serum, exhibits weaker androgen activity. A case-control study on ovarian cancer has revealed that elevated androsterone glucuronide levels lead to an increased risk of non-serious ovarian cancer in women [62]. Similarly, another study conducted by Kalogera et al [61, 63] also found significantly increased androsterone glucuronide levels in patients with lobular neoplasia compared to those with benign breast disease. Thus, androsterone glucuronide could be a valuable marker for assessing the efficacy of endocrine therapy for breast cancer by detecting its concentration in the blood and evaluating total androgen activity in an accurate way [63]. Some studies have shown a weak but positive correlation between the levels of androsterone glucuronide and the risk of prostate cancer [61]. Interestingly, Chinese men have lower plasm levels of androsterone glucuronide compared to Caucasian males in Western countries [64]. These studies suggest a positive correlation between androsterone glucuronide and the risk of hormone-dependent malignant tumours. However, the relationship between androsterone glucuronide and the risk of non-hormone-dependent malignant tumours remains unclear. In our study, we found that the increase in bilirubin (Z, Z) to androsterone glucuronide ratio is a protective factor for UGI cancers. This protective effect is primarily attributed to the anticancer effect of bilirubin. Given the uncertain relationship between androsterone glucuronide and non-hormone-dependent malignant tumours, it is possible that androsterone glucuronide contributes to the risk of EC and GC. However, bilirubin's anticancer effects likely outweigh any potential risks posed by androsterone glucuronide. The synergistic effect of bilirubin’s anticancer properties and the potential protective role of androsterone glucuronide suggests that an increased bilirubin (Z, Z) to androsterone glucuronide ratio may improve the prognosis of EC and GC. Further research is necessary to elucidate the specific underlying mechanisms.
Limitations
This study is subject to several limitations. First, the metabolomic profiles were derived from non-fasting plasma samples. Although corrections were made considering the time since last eating or drinking, residual variances that have not been accounted for could remain. Second, the investigation concentrated on gene-metabolite associations that are currently supported by gene expression data and biological knowledge, with a particular emphasis on those involving effector genes. This approach may overlook the potential significance of other metabolites or metabolic ratios that, despite their strong heritability, were not the primary focus of this study. To uncover the effector genes corresponding to these additional metabolites and ratios, future studies will need to integrate more comprehensive gene expression data and a deeper understanding of metabolic pathways. Third, our study's implementation of MR was limited by the fact that the majority of metabolites and their corresponding ratios were linked to only one IV. This restriction prevented us from using several sensitivity analyses, such as MR-Egger regression, which requires multiple IVs for robustness checks against potential biases like horizontal pleiotropy. Nevertheless, we sought to minimize the impact of horizontal pleiotropy by selecting IVs that are closely associated with specific effector genes known to directly influence metabolite concentrations. We also manually examined instances of metabolic pleiotropy and excluded IVs that affected multiple metabolites not part of the same biological pathway. Although these measures aimed to curtail bias, we recognize that it cannot be entirely ruled out due to current limitations in metabolomic profiling and incomplete data on metabolite-protein interactions. Future studies with a more exhaustive analysis of the metabolome will be vital for a more precise delineation of genetic impacts on metabolites.
Moreover, the demographic scope of this study was largely confined to older adults of European ancestry. Broadening the research to include a wider range of ages and ethnic backgrounds will be instrumental in validating and extending the applicability of our findings regarding genetic variations in metabolites and their ratios across different populations. Furthermore, our study lacks SNP, using a single SNP as an instrument in MR studies can lead to weak instrument bias, where the instrument is not sufficiently associated with the exposure, leading to imprecise estimates and potentially biased results.
Conclusion
In this MR study, we scrutinized the causal links between metabolite ratios and the risk of UGI cancers. Our rigorous analysis identified two critical metabolic ratios, the phosphate-to-alanine ratio and the bilirubin-to-androsterone glucuronide ratio, as significant players in the context of UGI cancers. The results suggest a potential risk relationship with an increased phosphate-to-alanine ratio, hinting at disruptions in energy metabolism and amino acid balance that may contribute to cancer development. Conversely, a higher bilirubin-to-androsterone glucuronide ratio appears to be associated with a reduced risk of UGI cancers, possibly reflecting the protective antioxidative effects of bilirubin and the influence of steroid hormone metabolism. These findings provide a promising step forward in understanding the metabolic underpinnings of UGI cancers and highlight the utility of MR in uncovering potential biomarkers for disease risk. While these results are encouraging, further investigation is essential to confirm these relationships and to explore their clinical implications. Ultimately, understanding these hub metabolites could lead to improved strategies for prevention, early detection and targeted treatment of UGI cancers.
Conflicts of interest
The authors declare no competing interests.
Funding
This study was supported by the Natural Science Foundation of Guangdong Province (Grant No. 2023A1515012548).
Consent for publication
All the authors of the article agreed to be published in the journal.
Ethics approval
Not applicable.
Availablity of data and material
All the data for this article can be found on GWAS database and UKbiobank database.
Author contributions
Pengkhun Nov collect data, analysing, interpretation of data, Duanyu Wang drafting the article; Wandan Li choose the topic; Kunpeng Du and Jiqiang Li designing, revising and guiding the study. The authors read and approved.
References
1. Pennathur A, Gibson MK, and Jobe BA, et al (2013) Oesophageal carcinoma Lancet 381 400–412 https://doi.org/10.1016/S0140-6736(12)60643-6 PMID: 23374478
2. Zong L, Abe M, and Seto Y, et al (2016) The challenge of screening for early gastric cancer in China Lancet 388 2606 https://doi.org/10.1016/S0140-6736(16)32226-7 PMID: 27894662
3. Sung H, Ferlay J, and Siegel RL, et al (2021) Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries CA Cancer J Clin 71(3) 209–249 https://doi.org/10.3322/caac.21660 PMID: 33538338
4. Huang S, Guo Y, and Li Z, et al (2020) A systematic review of metabolomic profiling of gastric cancer and esophageal cancer Cancer Biol Med 17(1) 181–198 https://doi.org/10.20892/j.issn.2095-3941.2019.0348 PMID: 32296585 PMCID: 7142846
5. Abbassi-Ghadi N, Kumar S, and Huang J, et al (2013) Metabolomic profiling of oesophago-gastric cancer: a systematic review Eur J Cancer 49 3625–3637 https://doi.org/10.1016/j.ejca.2013.07.004 PMID: 23896378
6. Gu J, Hu X, and Shao W, et al (2016) Metabolomic analysis reveals altered metabolic pathways in a rat model of gastric carcinogenesis Oncotarget 7 60053–60073 https://doi.org/10.18632/oncotarget.11049 PMID: 27527852 PMCID: 5312368
7. Kim KB, Yang JY, and Kwack SJ, et al (2010) Toxicometabolomics of urinary biomarkers for human gastric cancer in a mouse model J Toxicol Environ Health A 73 1420–1430 https://doi.org/10.1080/15287394.2010.511545 PMID: 20954069
8. Matsunaga S, Nishiumi S, and Tagawa R, et al (2018) Alterations in metabolic pathways in gastric epithelial cells infected with Helicobacter pylori Microb Pathog 124 122–129 https://doi.org/10.1016/j.micpath.2018.08.033 PMID: 9. Di Gialleonardo V, Tee SS, and Aldeborgh HN, et al (2016) High-throughput indirect quantitation of 13C enriched metabolites using 1H NMR Anal Chem 88 11147–11153 https://doi.org/10.1021/acs.analchem.6b03307 PMID: 27749041 PMCID: 5148664
10. Holmes E, Wilson ID, and Nicholson JK (2008) Metabolic phenotyping in health and disease Cell 134 714–717 https://doi.org/10.1016/j.cell.2008.08.026 PMID: 18775301
11. Wishart DS (2016) Emerging applications of metabolomics in drug discovery and precision medicine Nat Rev Drug Discov 15 473–484 https://doi.org/10.1038/nrd.2016.32 PMID: 26965202
12. Chan AW, Gill RS, and Schiller D, et al (2014) Potential role of metabolomics in diagnosis and surveillance of gastric cancer World J Gastroenterol 20 12874–12882 https://doi.org/10.3748/wjg.v20.i36.12874 PMID: 25278684 PMCID: 4177469
13. Xiao S and Zhou L (2017) Gastric cancer: metabolic and metabolomics perspectives (Review) Int J Oncol 51 5–17 https://doi.org/10.3892/ijo.2017.4000 PMID: 28535006
14. Yuan LW, Yamashita H, and Seto Y (2016) Glucose metabolism in gastric cancer: the cutting-edge World J Gastroenterol 22 2046–2059 https://doi.org/10.3748/wjg.v22.i6.2046 PMID: 26877609 PMCID: 4726677
15. Jayavelu ND and Bar NS (2014) Metabolomic studies of human gastric cancer: review World J Gastroenterol 20 8092–8101 https://doi.org/10.3748/wjg.v20.i25.8092 PMID: 25009381 PMCID: 4081680
16. Smith GD and Ebrahim S (2003) 'Mendelian randomization': can genetic epidemiology contribute to understanding environmental determinants of disease? Int J Epidemiol 32(1) 1–22 https://doi.org/10.1093/ije/dyg070 PMID: 12689998
17. Davey Smith G and Hemani G (2014) Mendelian randomization: genetic anchors for causal inference in epidemiological studies Hum Mol Genet 23(R1) R89–R98 https://doi.org/10.1093/hmg/ddu328 PMID: 25064373 PMCID: 4170722
18. Chen Y, Lu T, and Pettersson-Kymmer U, et al (2023) Genomic atlas of the plasma metabolome prioritizes metabolites implicated in human diseases Nat Genet 55(1) 44–53 https://doi.org/10.1038/s41588-022-01270-1 PMID: 36635386 PMCID: 7614162
19. Suhre K, Shin SY, and Petersen AK, et al (2011) Human metabolic individuality in biomedical and pharmaceutical research Nature 477 54–60 https://doi.org/10.1038/nature10354 PMID: 21886157 PMCID: 3832838
20. Sidore C, Busonero F, and Maschio A, et al (2015) Genome sequencing elucidates sardinian genetic architecture and augments association analyses for lipid and blood inflammatory markers Nat Genet 47(11) 1272–1281 https://doi.org/10.1038/ng.3368 PMID: 26366554 PMCID: 4627508
21. Sun Y, Zhou J, and Ye K (2021) White blood cells and severe COVID-19: a mendelian randomization study J Pers Med 11(3) 95 https://doi.org/10.3390/jpm11030195
22. Cai J, Li X, and Wu S, et al (2022) Assessing the causal association between human blood metabolites and the risk of epilepsy J Transl Med 20(1) 437 https://doi.org/10.1186/s12967-022-03648-5 PMID: 36180952 PMCID: 9524049
23. Zeng P, Wang T, and Zheng J, et al (2019) Causal association of type 2 diabetes with amyotrophic lateral sclerosis: new evidence from mendelian randomization using GWAS summary statistics BMC Med 17(1) 225 https://doi.org/10.1186/s12916-019-1448-9 PMID: 31796040 PMCID: 6892209
24. Burgess S, Small DS, and Thompson SG (2017) A review of instrumental variable estimators for mendelian randomization Stat Methods Med Res 26(5) 2333–2355 https://doi.org/10.1177/0962280215597579 PMCID: 5642006
25. Bowden J, Smith GD, and Haycock PC, et al (2016) Consistent estimation in mendelian randomization with some Invalid Instruments using a weighted median estimator Genet Epidemiol 40(4) 304–314 https://doi.org/10.1002/gepi.21965 PMID: 27061298 PMCID: 4849733
26. Hartwig FP, Davey Smith G, and Bowden J (2017) Robust inference in summary data mendelian randomization via the zero modal pleiotropy assumption Int J Epidemiol 46(6) 1985–1998 https://doi.org/10.1093/ije/dyx102 PMID: 29040600 PMCID: 5837715
27. Yavorska OO and Burgess S (2017) MendelianRandomization: an R package for performing mendelian randomization analyses using summarized data Int J Epidemiol 46(6) 1734–1739 https://doi.org/10.1093/ije/dyx034 PMID: 28398548 PMCID: 5510723
28. Verbanck M, Chen CY, and Neale B, et al (2018) Detection of widespread horizontal pleiotropy in causal relationships inferred from mendelian randomization between complex traits and diseases Nat Genet 50(5) 693 https://doi.org/10.1038/s41588-018-0099-7 PMID: 29686387 PMCID: 6083837
29. Warburg O, Wind F, and Negelein E (1927) The metabolism of tumors in the body J Gen Physiol 8(6) 519–530 https://doi.org/10.1085/jgp.8.6.519 PMID: 19872213 PMCID: 2140820
30. Jin HR, Wang J, and Wang ZJ, et al (2023) Lipid metabolic reprogramming in tumor microenvironment: from mechanisms to therapeutics J Hematol Oncol 16(1) 103 https://doi.org/10.1186/s13045-023-01498-2 PMID: 37700339 PMCID: 10498649
31. Zhu J, Shu X, and Guo X, et al (2020) Associations between genetically predicted blood protein biomarkers and pancreatic cancer risk Cancer Epidemiol Biomarkers Prev 29(7) 1501–1508 https://doi.org/10.1158/1055-9965.EPI-20-0091 PMID: 32439797 PMCID: 7334065
32. Tuerhong A, Xu J, and Shi S, et al (2021) Overcoming chemoresistance by targeting reprogrammed metabolism: the Achilles' heel of pancreatic ductal adenocarcinoma Cell Mol Life Sci 78(14) 5505–5526 https://doi.org/10.1007/s00018-021-03866-y PMID: 34131808 PMCID: 11072422
33. Kambakamba P, Schneider MA, and Linecker M, et al (2023) Early postoperative serum phosphate drop predicts sufficient hypertrophy after liver surgery Ann Surg 278(5) 763–771 https://doi.org/10.1097/SLA.0000000000006013 PMID: 37465990
34. Kato H, Kimura S, and Taguchi M, et al (2023) FGF23-related hypophosphatemia in a patient with small cell lung cancer: a case report and literature review Endocr J 70(10) 1005–1013 https://doi.org/10.1507/endocrj.EJ23-0086 PMID: 37468265
35. Vučetić M, Cormerais Y, and Parks S K, et al (2017) The central role of amino acids in cancer Redox homeostasis: vulnerability points of the cancer redox code Front Oncol 7 319 https://doi.org/10.3389/fonc.2017.00319
36. Ludwig K, Belle JEL, and Muthukrishnan SD, et al (2023) Nicotinamide adenine dinucleotide phosphate oxidase promotes glioblastoma radiation resistance in a phosphate and tensin homolog-dependent manner Antioxid Redox Signal 39(13-15) 890–903 https://doi.org/10.1089/ars.2022.0086 PMID: 37470216 PMCID: 10775910
37. Ran M, Zhou Y, and Guo Y, et al (2023) Cytosolic malic enzyme and glucose-6-phosphate dehydrogenase modulate redox balance in NSCLC with acquired drug resistance FEBS J 290(19) 4792–4809 https://doi.org/10.1111/febs.16897 PMID: 37410361
38. Mohelnikova-Duchonova B, Marsakova L, and Vrana D, et al (2011) Superoxide dismutase and nicotinamide adenine dinucleotide phosphate: quinone oxidoreductase polymorphisms and pancreatic cancer risk Pancreas 40(1) 72–78 https://doi.org/10.1097/MPA.0b013e3181f74ad7
39. Shi H, Cheng Y, and Shi Q, et al (2022) Myoferlin disturbs redox equilibrium to accelerate gastric cancer migration Front Oncol 12 905230 https://doi.org/10.3389/fonc.2022.905230 PMID: 36147922 PMCID: 9486956
40. Vettore L, Westbrook RL, and Tennant DA (2020) New aspects of amino acid metabolism in cancer Br J Cancer 122(2) 150–156 https://doi.org/10.1038/s41416-019-0620-5 PMCID: 7052246
41. Groen AK, Sips HJ, and Vervoorn RC, et al (1982) Intracellular compartmentation and control of alanine metabolism in rat liver parenchymal cells Eur J Biochem 122(1) 87–93 https://doi.org/10.1111/j.1432-1033.1982.tb05851.x PMID: 7060572
42. He MJ, Pu W, and Wang X, et al (2022) Spatial metabolomics on liver cirrhosis to hepatocellular carcinoma progression Cancer Cell Int 22(1) 366 https://doi.org/10.1186/s12935-022-02775-9 PMID: 36419080 PMCID: 9686114
43. Budczies J, Brockmöller SF, and Müller BM, et al (2013) Comparative metabolomics of estrogen receptor positive and estrogen receptor negative breast cancer: alterations in glutamine and beta-alanine metabolism J Proteomics 94 279–288 https://doi.org/10.1016/j.jprot.2013.10.002 PMID: 24125731
44. Elia I, Rossi M, and Stegen S, et al (2019) Breast cancer cells rely on environmental pyruvate to shape the metastatic niche Nature 568(7750) 117–121 https://doi.org/10.1038/s41586-019-0977-x PMID: 30814728 PMCID: 6451642
45. Vogel ME and Zucker SD (2016) Bilirubin acts as an endogenous regulator of inflammation by disrupting adhesion molecule-mediated leukocyte migration Inflamm Cell Signal 3(1) e1178 PMID: 26925435 PMCID: 4768809
46. Tomaro ML and Batlle AM (2002) Bilirubin: its role in cytoprotection against oxidative stress Int J Biochem Cell Biol 34(3) 216–220 https://doi.org/10.1016/S1357-2725(01)00130-3 PMID: 11849989
47. Doumas BT and Wu TW (1991) The measurement of bilirubin fractions in serum Crit Rev Clin Lab Sci 28(5-6) 415–445 https://doi.org/10.3109/10408369109106872 PMID: 1772588
48. Li N, Xu M, and Cai MY, et al (2015) Elevated serum bilirubin levels are associated with improved survival in patients with curatively resected non-small-cell lung cancer Cancer Epidemiol 39(5) 763–768 https://doi.org/10.1016/j.canep.2015.06.007 PMID: 26159684
49. Liu X, Meng QH, and Ye Y, et al (2015) Prognostic significance of pretreatment serum levels of albumin, LDH and total bilirubin in patients with non-metastatic breast cancer Carcinogenesis 36(2) 243–248 https://doi.org/10.1093/carcin/bgu247
50. Ioannou GN, Liou IW, and Weiss NS (2006) Serum bilirubin and colorectal cancer risk: a population-based cohort study Aliment Pharmacol Ther 23(11) 1637–1642 https://doi.org/10.1111/j.1365-2036.2006.02939.x PMID: 16696814
51. Wei TT, Wang LL, and Yin JR, et al (2017) Relationship between red blood cell distribution width, bilirubin, and clinical characteristics of patients with gastric cancer Int J Lab Hematol 39(5) 497–501 https://doi.org/10.1111/ijlh.12675 PMID: 28497572
52. Huang X, Chen Y, and Yang H, et al (2023) Preoperative serum bilirubin is an independent prognostic factor for curatively resected esophageal squamous cell carcinoma BMC Cancer 23(1) 706 https://doi.org/10.1186/s12885-023-11215-4 PMID: 37507653 PMCID: 10375695
53. Fuhrman MP, Charney P, and Mueller CM (2004) Hepatic proteins and nutrition assessment J Am Diet Assoc 104(8) 1258–1264 https://doi.org/10.1016/j.jada.2004.05.213 PMID: 15281044
54. Szor DJ, Pereira MA, and Ramos M, et al (2023) Preoperative albumin-bilirubin score is a prognostic factor for gastric cancer patients after curative gastrectomy World J Gastrointest Surg 15(6) 1125–1137 https://doi.org/10.4240/wjgs.v15.i6.1125 PMID: 37405095 PMCID: 10315126
55. Yagyu T, Saito H, and Sakamoto T, et al (2019) Preoperative albumin-bilirubin grade as a useful prognostic indicator in patients with pancreatic cancer Anticancer Res 39(3) 1441–1446 https://doi.org/10.21873/anticanres.13260 PMID: 30842180
56. Lee HG, Lim SB, and Lee JL, et al (2022) Preoperative albumin-bilirubin score as a prognostic indicator in patients with stage III colon cancer Sci Rep 12(1) 14910 https://doi.org/10.1038/s41598-022-19329-8 PMID: 36050367 PMCID: 9437055
57. Kinoshita F, Yamashita T, and Oku Y, et al (2021) Prognostic impact of albumin-bilirubin (ALBI) grade on non-small lung cell carcinoma: a propensity-score matched analysis Anticancer Res 41(3) 1621–1628 https://doi.org/10.21873/anticanres.14924 PMID: 33788758
58. Aoyama T, Ju M, and Machida D, et al (2022) Clinical impact of preoperative albumin-bilirubin status in esophageal cancer patients who receive curative treatment In Vivo 36(3) 1424–1431 https://doi.org/10.21873/invivo.12847 PMID: 35478112 PMCID: 9087095
59. Luo X, Chen XD, and Chen JL, et al (2023) High albumin-bilirubin grade predicts worse short-term complications in gastric cancer patients with metabolic syndrome: a retrospective study J Gastrointest Oncol 14(5) 2039–2047 https://doi.org/10.21037/jgo-23-599 PMID: 37969839 PMCID: 10643593
60. Zhang X J, Fei H, and Sun CY, et al (2023) Novel prognostic score based on the preoperative total bilirubin-albumin ratio and fibrinogen-albumin ratio in ampullary adenocarcinoma World J Gastrointest Surg 15(10) 2247–2258 https://doi.org/10.4240/wjgs.v15.i10.2247 PMID: 37969714 PMCID: 10642462
61. Kalogera E, Pistos C, and Provatopoulou X, et al (2013) Androgen glucuronides analysis by liquid chromatography tandem-mass spectrometry: could it raise new perspectives in the diagnostic field of hormone-dependent malignancies? J Chromatogr B Analyt Technol Biomed Life Sci 940 24–34 https://doi.org/10.1016/j.jchromb.2013.09.022 PMID: 24140653
62. Trabert B, Michels KA, and Anderson GL, et al (2019) Circulating androgens and postmenopausal ovarian cancer risk in the women's health initiative observational study Int J Cancer 145(8) 2051–2060 https://doi.org/10.1002/ijc.32157 PMID: 30684389 PMCID: 6660427
63. Kalogera E, Pistos C, and Provatopoulou X, et al (2016) Bioanalytical LC-MS method for the quantification of plasma androgens and androgen glucuronides in breast cancer J Chromatogr Sci 54(4) 583–592 https://doi.org/10.1093/chromsci/bmv190 PMID: 26762957 PMCID: 4885394
64. Santner SJ, Albertson B, and Zhang GY, et al (1998) Comparative rates of androgen production and metabolism in Caucasian and Chinese subjects J Clin Endocrinol Metab 83(6) 2104–2109 PMID: 9626146
Supplementary material
Supplementary Table 1. The pleiotropy of causal relationship between all metabolites and esophageal cancer.
Supplementary Table 2. The heterogeneity of causal relationship between all metabolites and esophageal cancer.
Supplementary Table 3. The pleiotropy of causal relationship between all metabolites and gastric cancer.
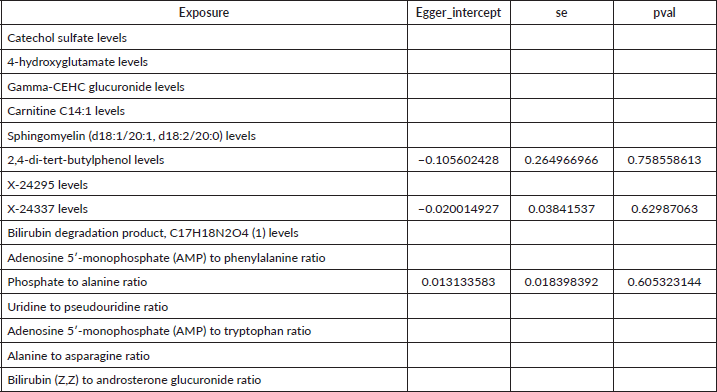
Supplementary Table 4. The heterogeneity of causal relationship between all metabolites and gastric cancer.
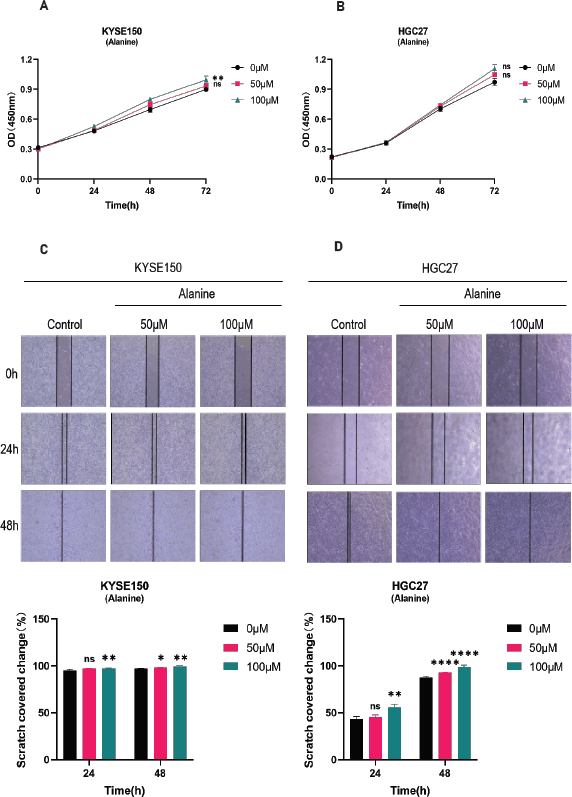
Supplementary Figure 1. A-B: CCK-8 method was used to detect the proliferation of KYSE150 and HGC27 cells. Divide different cell lines into three groups and treat the cells with different concentrations of alanine (0, 50, 100 μ M). Measure the absorbance values of cells at different time points (0, 24, 48, 72 hours) using the CCK-8 method, and evaluate the proliferation of cells through plotting. C-D: Scratch assay for detecting the migration of KYSE150 and HGC27 cells. KYSE150 and HGC27 cells were treated with different concentrations of alanine for 24 and 48 hours, and the scratch area at different time points was calculated to evaluate the migration ability of the cells. All experiments were repeated 3 times, and the results were expressed as mean ± SDs. Compared with the control group, there was no significant difference in ns, with * * p < 0.05, p < 0.01, * * * p < 0.005, * * * * < 0.001