Comprehensive germline profiling of patients with breast cancer: initial experience from a Familial Cancer Clinic
Raja Pramanik1, Sindhura Chitikela1, S V S Deo2, Ajay Gogia1, Atul Batra1, Akash Kumar3, Ritu Gupta4, Deepshi Thakral4, Vedam L Ramprasad5, Sandeep Mathur6, D N Sharma7, Aparna Sharma1, Ashutosh Mishra2 and Babul Bansal2
1Department of Medical Oncology, All India Institute of Medical Sciences, New Delhi 110029, India
2Department of Surgical Oncology, All India Institute of Medical Sciences, New Delhi 110029, India
3Department of Medical Oncology, National Cancer Institute, NCI Jhajjar 124105, India
4Department of Laboratory Oncology, All India Institute of Medical Sciences, New Delhi 110029, India
5Medgenome Labs Ltd, Bengaluru 560099, India
6Department of Pathology, All India Institute of Medical Sciences, New Delhi 110029, India
7Department of Radiation Oncology, All India Institute of Medical Sciences, New Delhi 110029, India
ahttps://orcid.org/0000-0003-0483-6254
Abstract
Introduction: Breast cancer is the most common cancer among Indian females. There is limited data on germline profiling of breast cancer patients from India.
Objective: The objective of the current study was to analyse the frequency and spectrum of germline variant profiles and clinicopathological characteristics of breast cancer patients referred to our Familial Cancer Clinic (FCC).
Materials and methods: It is a single-centre audit of patients with a confirmed diagnosis of breast carcinoma referred to our FCC from January 2017 to 2020. All patients underwent pretest counselling. Genetic testing was done by multigene panel testing by next-generation sequencing along with reflex multiplication ligation-dependent probe amplification for BRCA1 and 2. The variants were classified based on American College of Medical Genetics guidelines. Demographic and clinicopathological details were extracted from the case record files.
Results: One hundred and fifty-five patients were referred to the FCC and underwent pretest counselling. A total of 99 (63.9%) patients underwent genetic testing. Among them, 62 patients (62/99 = 62.6%) had a germline variant. A pathogenic/likely pathogenic (P/LP) germline variant was identified in 41 (41.4%) of the patients who underwent testing. Additional variants of unknown significance (VUS) were identified in seven patients who also carried a P/LP variant. VUS alone was detected in 21 patients (21/99 = 21.2%). Among the P/LP pathogenic variants (PV), BRCA 1 PV were seen in 27 patients (65.8%), BRCA 2 variants in 7 patients (17.1%), ATM variants in 3 patients (7.3%) and RAD51, TP53, CHEK2 and HMMR in 1 patient each. Variants were significantly more common in patients with a family history (FH) of malignancy than those without FH (58.5% versus 29.5%; p = 0.013). Age and triple-negative histology were not found to be significantly associated with the occurrence of P/LP PVs.
Conclusion: We report a 41% P/LP variant rate in our selected cohort of breast cancer patients, with variants in BRCA constituting 83% and non-BRCA gene variants constituting 17%.
Keywords: germline variant, next-generation sequencing, breast cancer, India
Correspondence to: Raja Pramanik
Email: drrajapramanik@gmail.com
Published: 15/02/2024
Received: 11/10/2023
Publication costs for this article were supported by ecancer (UK Charity number 1176307).
Copyright: © the authors; licensee ecancermedicalscience. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Introduction
Breast cancer is the most common cancer representing 11.7% of cancers worldwide and the most common cause of cancer mortality in females [1]. It is the most common cancer among females in India [1, 2]. Genetic predisposition is the most important risk factor for the occurrence of breast cancer [3]. Approximately 10%–15% of all breast cancers can be attributed to the presence of germline pathogenic variants (PVs) involving BRCA and non-BRCA breast cancer susceptibility genes. Variants in BRCA 1 and 2 account for up to 50% of all familial breast cancer [4–6]. Other high-risk genes include PALB2, CDH1, PTEN, TP53 and STK11 with a lifetime risk of breast cancer of up to 50%–80%. Variants involving ATM, CHEK2, BRIP1, RAD51C, RAD51D, NBN and BARD1 are known to be associated with a moderately increased risk of breast cancer and a lifetime risk of around 25% [7, 8]. Testing and identification of these PVs is important to identify high-risk individuals, screening for early diagnosis of breast and/or ovarian cancer and accordingly plan for risk reduction and therapeutic strategies. There is limited literature on germline profiling in breast cancer cases, reported from India.
We report the results of a retrospective audit aimed at analysing the genetic and clinicopathologic profile of patients with breast cancer referred to a Familial Cancer Clinic (FCC) at a tertiary care cancer centre in India.
Methods
This retrospective observational study was carried out at a tertiary care centre in India. Patients with histopathological diagnosis of carcinoma breast who were referred to our genetic clinic between January 2017 and 2020 were analysed in the study. Pre-test counselling and post-test counselling were done by a team of medical oncologists and surgical oncologists. The genetic test performed included multigene panel testing by next-generation sequencing (NGS) along with reflex multiplication ligation-dependent probe amplification (MLPA) to detect large deletions/duplications in BRCA1 and 2. Those with pathogenic/likely pathogenic (P/LP) variants in cancer predisposition genes were offered post-test counselling and further management was formulated in the multidisciplinary tumour board. The variants were classified based on American College of Medical Genetics (ACMG) guidelines.
Genetic testing
Briefly, blood samples (8 mL) were collected from each patient and DNA extraction was done. A targeted gene panel of 104 genes (listed in Supplementary Table S1) was performed using a custom capture kit. Gene libraries were sequenced to mean > 80–100× coverage on the Illumina sequencing platform. Sequences were aligned to the human reference genome (GRCh38.p13). Gene annotation of the variants was performed using Ensembl Variant Effect Predictor programme against the Ensembl release 99 human gene model. Reflex MLPA testing was done for patients with no identifiable germline variants. Copy number variations in 24 exons of BRCA 1 and 27 exons of BRCA 2 were identified by hybridising with MLPA assay. Each MLPA probe consisted of two hemi-probes bound to an adjoining site of the target sequence. Upon ligation and PCR amplification, an amplicon of a unique length was generated by each probe. Copy number changes of different exons between test and control DNA samples were analysed by detecting the MLPA peak pattern. Variants identified were classified as pathogenic (class 5), likely pathogenic (class 4), variants of unknown significance/VUS (class 3), likely benign (class 2) and benign (class 1) according to ACMG guidelines. All P/LP variants were reported using the Human Genome Variant Society (HGVS) nomenclature and the most frequent transcripts. For example, NM_7297.4 (7,088 bp) and NP_009225.1 (1,863 amino acids) for BRCA1 germline variants.
Data regarding demographic characteristics, clinical profile, histopathology and family history (FH) were extracted from the files in the medical record system.
The primary outcome was the frequency of P/LP PVs in the cohort. Secondary outcomes included the identification of clinicopathological features, their association and the spectrum of PVs.
Statistics
Categorical data were represented using descriptive statistics in the form of percentages, medians and ranges. Numerical data were represented as median and range. Chi-square tests and Fisher’s exact tests were used to compare and examine the relationship between variables. All tests of the hypothesis were conducted at an alpha level of 0.05, with a 95% confidence interval. Statistical analysis was performed using IBM SPSS version 26 software.
Results
A total of 155 patients were referred to our genetic clinic during this period. Most of the patients (n = 151) were referred from the breast cancer clinic of AIIMS New Delhi or NCI-AIIMS, Jhajjar. Four patients were referred from the oncology clinics of Army Hospital (R&R) in New Delhi. The median age of the entire cohort was 42 years (range 21–80 years). All patients were female except one.
Pre-test counselling was done for all the patients and all 155 qualified the contemporary National Comprehensive Cancer Network (NCCN) criteria for genetic testing. All 155 patients were advised genetic testing by a multigene NGS panel. Ethnicity was assessed by their religion and place of residence. There were 129 (83.22%) Hindus, 8 (5.16%) Muslims, 6 (3.87%) Christians and 11 (7.09%) Sikhs. Overall, 59% of patients were from urban areas and 58% were literate.
Eight patients had bilateral breast cancer with synchronous breast cancer in three of them, while seven patients had both breast and ovarian cancer (Table 1).
Information on FH was available for 107 patients. A FH of breast and/or ovarian cancer in first-degree relatives was present in 35 patients (32.71%). A FH of malignancies other than breast and ovarian cancer was seen in 11 patients (10.28%). The FH of malignancy in second-degree relatives was present in a total of 42 patients (39.25%) (Table 1).
A total of 99 (63.9%) patients underwent genetic testing. The reasons for not undergoing testing included financial issues (50/56 = 89%) and patient preference (6/50 = 11%). Of these 99, 62 patients (62/99 = 62.6%) had a germline variant. As a whole, 73 variants were identified in 62 patients. Of those with variants, most of the patients (52/62 (87%)) had a single variant, with two variants in nine (14.5%) and three variants in one patient (1.6%), respectively (Table 2).
A P/LP variant was identified in 41 (41/99 = 41.4%) of the patients who underwent testing. Additional VUS were identified in seven patients who also carried a P/LP variant. VUS alone were detected in 21 patients (21/99 = 21.2%). Two patients had VUS involving 2 genes and 1 patient had a VUS each in 3 genes accounting for a total VUS variants of 25 (Table 2 and Supplementary Table S2).
Clinical characteristics of patients with P/LP variants (n = 41)
The median age of patients with P/LP variants was 42 years. Nineteen patients (46.34%) belonged to the age group of less than or equal to 40 years. Information about FH was available for 31 patients amongst whom a FH of breast and/or ovarian cancer was observed in 24 patients (77.4%). Receptor status was available for 37 patients; amongst whom, triple-negative breast cancer (TNBC) was identified in 26 patients (70.3%), hormone-positive breast cancer in 9 patients (24.3%) and triple-positive breast cancer in 2 patients (5.4%) (Table 1, Supplementary Figure S2-S4).
Table 1. Baseline characteristics.
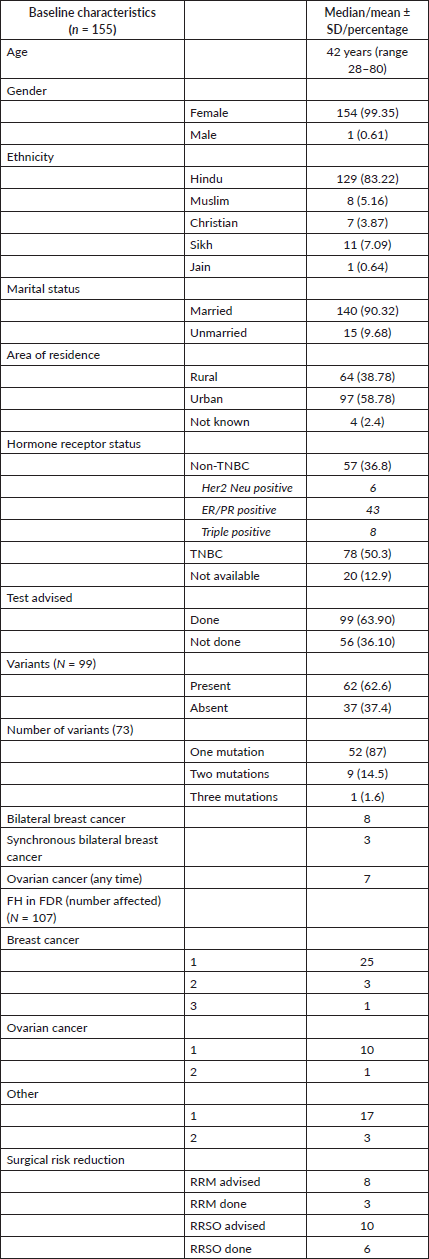
Table 2. Spectrum of variants.
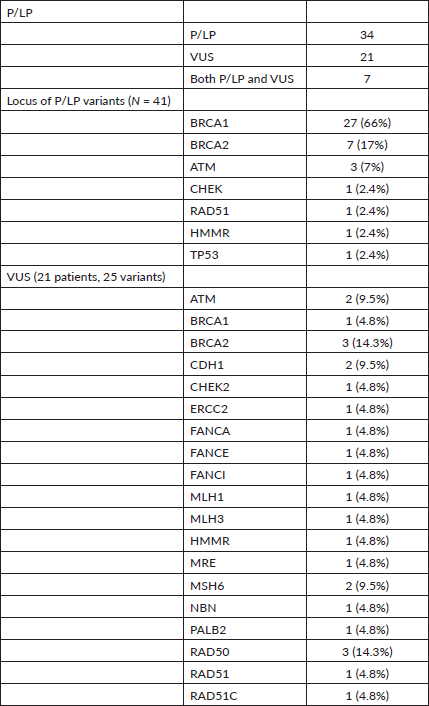
Among the P/LP variants, BRCA 1 variants were seen in 27 patients (65.8%), BRCA 2 variants in 7 patients (17.1%), ATM variants in 3 patients (7.3%) and RAD51, TP53, CHEK and HMMR in 1 patient each (Table 2, Supplementary Figure S1). The Ashkenazi Jews founder variant (c.68_69delAG) was seen in two patients. Large genomic rearrangement in BRCA1 were found in two patients (Supplementary Table S3, Figure 1).
Association of P/LP variants with FH, histology and age at diagnosis
Out of the 41 patients with a positive FH who underwent genetic testing, 24 patients had a P/LP variant (58.5%) compared to 7 patients among 27 patients who had no positive FH and underwent genetic testing (25.9%). Variants were significantly more common in patients with a FH of malignancy (p = 0.013) (Table 3).
Table 3. Association of P/LP mutation with age, histology and FH.
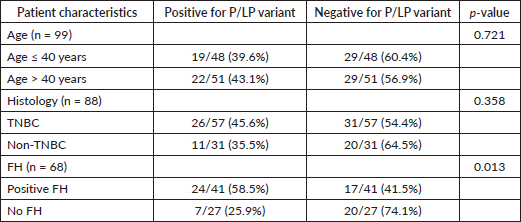
P/LP variants were present in 26 out of 57 tested patients with TNBC (45.6%). This was not significantly different from that of the non-TNBC cluster, in which P/LP variants were present in 11 out of 31 tested patients (35.48%) (p = 0.358) (Table 3). P/LP were present in 19 out of 48 patients tested in the age group ≤40 years (42.68%) and in 22 out of 51 patients tested in the age group >40 years (40%). The difference was not significant (p = 0.721) (Table 3).
Hereditary breast-ovarian cancer syndrome
Out of the seven patients who had both breast and ovarian cancer (metachronous), five had undergone genetic testing, with P/LP variant found in all five patients (four BRCA1 and 1 BRCA2). Out of the eight patients who had bilateral breast cancer, either synchronous or metachronous, six underwent genetic testing, out of which four had P/LP variants (three BRCA1, 1 TP53).
Discussion
The current study is an audit of patients with breast cancers who were selectively referred to our FCC over the initial 3 years of its functioning.
We report 41% positivity for P/LP PVs in this highly selected cohort of patients with carcinoma breast. BRCA 1 constituted the most mutated gene and accounted for 65%, followed by BRCA2 variants in 13.7% and non-BRCA genes constituted 22.6%. The prevalence of the Ashkenazi Jew founder variant was found to be 7%.
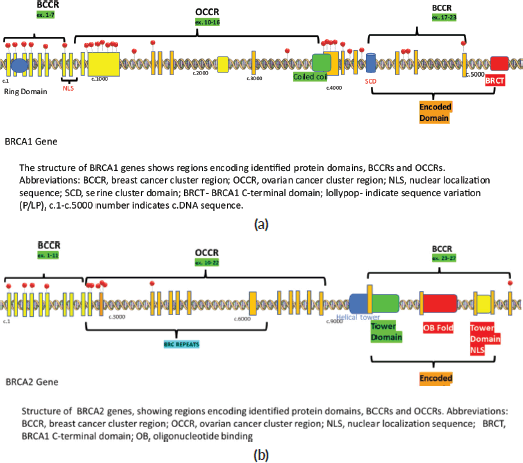
Figure 1. Lollipop plot of the detected P/LP variants during the study. (a): BRCA1 gene. (b): BRCA2 gene.
In a retrospective study by Singh et al [9], around 1,010 patients with breast and ovarian cancers were tested using a 14-gene panel, germline variants were identified in 30% of patients, with variants in BRCA genes constituting 84.9% and that of non-BRCA genes constituting 15.1%. They reported significantly higher variant detection rates in the age group of 40 years or younger, TNBC and in those with first-degree family members affected with breast and/or ovarian cancer [9]. Another study by Chheda et al [10] is a laboratory-based multicentre study of 160 unrelated women with breast and ovarian cancer or unaffected women with a FH. Variant testing was done for only BRCA 1 and BRCA 2 and they reported a frequency of 31.9% for P/LP PVs. A similar study from a tertiary care centre reported a comparable variant rate involving BRCA 1 in 22% and BRCA 2 in 8% [11]. Both the above-cited studies showed comparable variant detection rates to our study. However, there is a higher percentage of non-BRCA variants reported in our study 22.6% versus 15% in the study by Singh et al [9] and 11.9% in a study by Kadri et al [12] which could be attributed to the application of an extended targeted gene panel in our study. In a prospective study by Mittal et al [13], conducted at a tertiary centre in North India in which consecutive carcinoma breast patients (n = 236) were subjected to multigene hereditary germline panel testing, P/LP variants were found in 44/236 (18.64%) women; variants in BRCA1 (22/47, 46.8%) and BRCA2 (9/47, 19.1%) were the most common, with 34% of variants present in non-BRCA genes. This prevalence of 18.64% in unselected breast carcinoma patients may actually be close to the true prevalence of germline variants in the population. The variant detection rate is higher in our study because of the highly selected study population.
The prevalence of germline variants is significantly higher compared to that of reports from various ethnicities worldwide. A study by Peto et al [5] reported a prevalence of BRCA 1 and BRCA 2 of 3% in unselected breast cancer patients, respectively, in the UK with higher prevalence in those aged <36 years. A study by Tung et al [14] reported that among sequential patients with breast cancer (n = 488) from the USA, germline variants in cancer predisposition genes were found in 10.7%, using a panel of 25 predisposition genes including 6.1% in BRCA1/2 and 4.6% in other breast/ovarian cancer predisposition genes including CHEK2 (n = 10), ATM (n = 4), BRIP1 (n = 4) and one each in PALB2, PTEN, NBN, RAD51C, RAD51D, MSH6 and PMS2. BRCA variant rate was higher in patients of young age, of Ashkenazi Jewish ancestry, with TNBC and those with a FH of breast/ovarian cancer [14]. Another German study by Engel et al [15] on the prevalence of germline BRCA 1/2 variants in TNBC patients with no FH reported a variant prevalence of 15.8% in the overall cohort, 32.9% in the age group 20–29 years, significantly higher compared to 6.9% in the age group 60–69 years. In a Chinese study reported by Sun et al [6], 8,085 consecutive unselected breast cancer patients were enrolled, and germline variants were assessed using a 62-gene panel. P/LP PVs were identified in 9.2% of patients, 5.3% of patients had BRCA1 or BRCA2 variant (1.8% in BRCA1 and 3.5% in BRCA2) and 3.9% carried variants in non-BRCA genes with the highest prevalence in TNBCs. In a systematic review by Armstrong et al [16] of 70 studies, BRCA1/2 variant prevalence was reported to be around 1.8% in Spain to 15.4% in the United States, with a higher prevalence in TNBCs.
Our study is a retrospective analysis with a small sample of a highly selected group of patients preferentially referred for testing. The present study shows a high prevalence of variants in breast cancer predisposition genes in our setting and represents an audit of our initial experience of the FCC. The non-significant difference in the frequency of variants based on age and hormone status contrasts with the findings reported from other Indian studies as well as studies from other regions of the world [6, 9, 14, 15]. This may be attributed to the high selection and referral bias in our patient population.
With the advent of targeted therapies like poly (ADP-ribose) polymerase inhibitors in early as well as metastatic breast cancers, germline testing for breast cancer patients plays a pivotal role in the management of breast cancer [17–20]. NCCN [21] criteria for genetic testing have been substantially relaxed to the extent that almost every breast cancer patient merits a germline multi-gene panel testing.
The evidence quoted above may indicate a higher prevalence of germline variants in our setting compared to Western cohorts. Larger data using multi-gene panels in an unselected manner is the need of the hour to better qualify the exact prevalence and spectrum.
Variants in the South Asian population particularly India are less well represented in Western studies making variant calling difficult.
Conclusion
This initial experience from a newly established FCC at a tertiary cancer centre can serve as an example to understand practical and logistic real-world challenges in gathering data as well as help us in planning larger multicentre studies. Such studies are extremely relevant to assimilate India-specific data and knowledge about various variant and their clustering if any in the Indian subcontinent. Also, smaller sample sizes and referrals from clinics highlight the unmet need for clinician awareness in proactively pursuing genetic evaluations in breast cancer patients. Mainstreaming genetic testing into routine clinical care is the way forward to cater to our huge clinical burden.
Conflicts of interest
All the authors declare no potential conflict of interest.
Funding
Not applicable.
Ethical approval
The study protocol was approved by the Institute Ethics Committee vide letter number: IEC- 511/05.06.2020, RP-50/2020 dated 30.9.2020.
Consent to participate
Informed consent was obtained from all patients.
Author contributions
Authors, RP, SVSD, AG, AB, AK, AS, RG, DT, SM and VR contributed to the concept design and conduct of the study; RP, SC, BB, AM and AS did genetic counselling; and SC, AS and RP did the statistical analysis. SC and RP drafted the manuscript and all authors edited the manuscript.
Availability of data and material
Data regarding this study will be available from the corresponding author (RP) on reasonable request.
References
1. Sung H, Ferlay J, and Siegel RL, et al (2021) Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries CA Cancer J Clin 71(3) 209–249 https://doi.org/10.3322/caac.21660 PMID: 33538338
2. Mathur P, Sathishkumar K, and Chaturvedi M, et al (2020) Cancer statistics, 2020: report from National Cancer Registry Programme, India JCO Glob Oncol 6 1063–1075 https://doi.org/10.1200/GO.20.00122 PMID: 32673076 PMCID: 7392737
3. Claus EB, Risch N, and Thompson WD (1994) Autosomal dominant inheritance of early-onset breast cancer. Implications for risk prediction Cancer 73(3) 643–651 https://doi.org/10.1002/1097-0142(19940201)73:3<643::AID-CNCR2820730323>3.0.CO;2-5 PMID: 8299086
4. King MC, Marks JH, and Mandell JB, et al (2003) Breast and ovarian cancer risks due to inherited mutations in BRCA1 and BRCA2 Science 302(5645) 643–646 https://doi.org/10.1126/science.1088759 PMID: 14576434
5. Peto J, Collins N, and Barfoot R, et al (1999) Prevalence of BRCA1 and BRCA2 gene mutations in patients with early-onset breast cancer J Natl Cancer Inst 91(11) 943–949 https://doi.org/10.1093/jnci/91.11.943 PMID: 10359546
6. Sun J, Meng H, and Yao L, et al (2017) Germline mutations in cancer susceptibility genes in a large series of unselected breast cancer patients Clin Cancer Res 23(20) 6113–6119 https://doi.org/10.1158/1078-0432.CCR-16-3227 PMID: 28724667
7. Apostolou P and Fostira F (2013) Hereditary breast cancer: the era of new susceptibility genes BioMed Res Int 2013 e747318 https://doi.org/10.1155/2013/747318
8. Han MR, Zheng W, and Cai Q, et al (2017) Evaluating genetic variants associated with breast cancer risk in high and moderate-penetrance genes in Asians Carcinogenesis 38(5) 511–518 https://doi.org/10.1093/carcin/bgx010 PMID: 28419251 PMCID: 5963497
9. Singh J, Thota N, and Singh S, et al (2018) Screening of over 1000 Indian patients with breast and/or ovarian cancer with a multi-gene panel: prevalence of BRCA1/2 and non-BRCA mutations Breast Cancer Res Treat 170(1) 189–196 https://doi.org/10.1007/s10549-018-4726-x PMID: 29470806
10. Chheda P, Pande S, and Dama T, et al (2020) Spectrum of germline BRCA mutations in hereditary breast and ovarian cancer syndrome in Indian population: a central reference laboratory experience Cancer Res Stat Treat 3(1) 32 https://doi.org/10.4103/CRST.CRST_101_19
11. Mehta A, Vasudevan S, and Sharma SK, et al (2018) Germline BRCA1 and BRCA2 deleterious mutations and variants of unknown clinical significance associated with breast/ovarian cancer: a report from North India CMAR 10 6505–6516 https://doi.org/10.2147/CMAR.S186563
12. Kadri MSN, Patel KM, and Bhargava PA, et al (2021) Mutational landscape for Indian hereditary breast and ovarian cancer cohort suggests need for identifying population specific genes and biomarkers for screening Front Oncol 10 568786 https://doi.org/10.3389/fonc.2020.568786 PMID: 33552952 PMCID: 7859489
13. Mittal A, Deo SVS, and Gogia A, et al (2022) Profile of pathogenic mutations and evaluation of germline genetic testing criteria in consecutive breast cancer patients treated at a North Indian Tertiary Care Center Ann Surg Oncol 29(2) 1423–1432 https://doi.org/10.1245/s10434-021-10870-w
14. Tung N, Lin NU, and Kidd J, et al (2016) Frequency of germline mutations in 25 cancer susceptibility genes in a sequential series of patients with breast cancer J Clin Oncol 34(13) 1460–1468 https://doi.org/10.1200/JCO.2015.65.0747 PMID: 26976419 PMCID: 4872307
15. Engel C, Rhiem K, and Hahnen E, et al (2018) Prevalence of pathogenic BRCA1/2 germline mutations among 802 women with unilateral triple-negative breast cancer without family cancer history BMC Cancer 18(1) 265 https://doi.org/10.1186/s12885-018-4029-y PMID: 29514593 PMCID: 5842578
16. Armstrong N, Ryder S, and Forbes C, et al (2019) A systematic review of the international prevalence of BRCA mutation in breast cancer CLEP 11 543–561 https://doi.org/10.2147/CLEP.S206949
17. Tutt ANJ, Garber JE, and Kaufman B, et al (2021) Adjuvant olaparib for patients with BRCA1- or BRCA2-mutated breast cancer N Engl J Med 384 2394–2405 https://doi.org/10.1056/NEJMoa2105215 PMID: 34081848 PMCID: 9126186
18. Geyer CE Jr, Garber JE, and Gelber RD, et al (2022) Overall survival in the OlympiA phase III trial of adjuvant olaparib in patients with germline pathogenic variants in BRCA1/2 and high-risk, early breast cancer Ann Oncol 33 1250–1268 [https://www.annalsofoncology.org/article/S0923-7534(22)04165-5/fulltext] Date accessed: 21/02/23 https://doi.org/10.1016/j.annonc.2022.09.159 PMID: 36228963 PMCID: 10207856
19. Litton JK, Rugo HS, and Ettl J, et al (2018) Talazoparib in patients with advanced breast cancer and a germline BRCA mutation N Engl J Med 379(8) 753–763 https://doi.org/10.1056/NEJMoa1802905 PMID: 30110579 PMCID: 10600918
20. Litton JK, Hurvitz SA, and Mina LA, et al (2020) Talazoparib versus chemotherapy in patients with germline BRCA1/2-mutated HER2-negative advanced breast cancer: final overall survival results from the EMBRACA trial Ann Oncol 31 1526–1535 [https://www.annalsofoncology.org/article/S0923-7534(20)42106-4/fulltext] https://doi.org/10.1016/j.annonc.2020.08.2098 PMID: 32828825 PMCID: 10649377
21. NCCN Guidelines detail [https://www.nccn.org/guidelines/guidelines-detail] Date accessed: 21/02/23
Supplementary Table S1. List of genes in the targeted germline panel used during the study.

Supplementary Table S2. Patient-wise spectrum of VUS mutations.
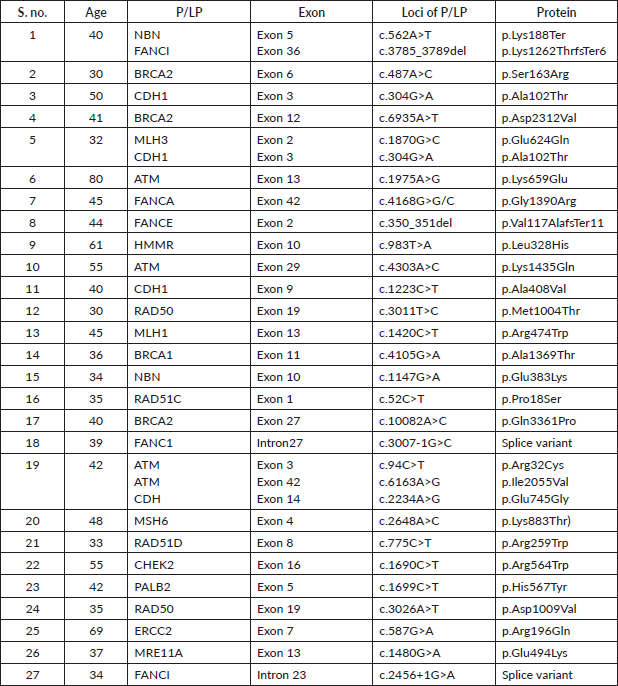
Supplementary Table S3a. P/LP variants in the BRCA1 gene.
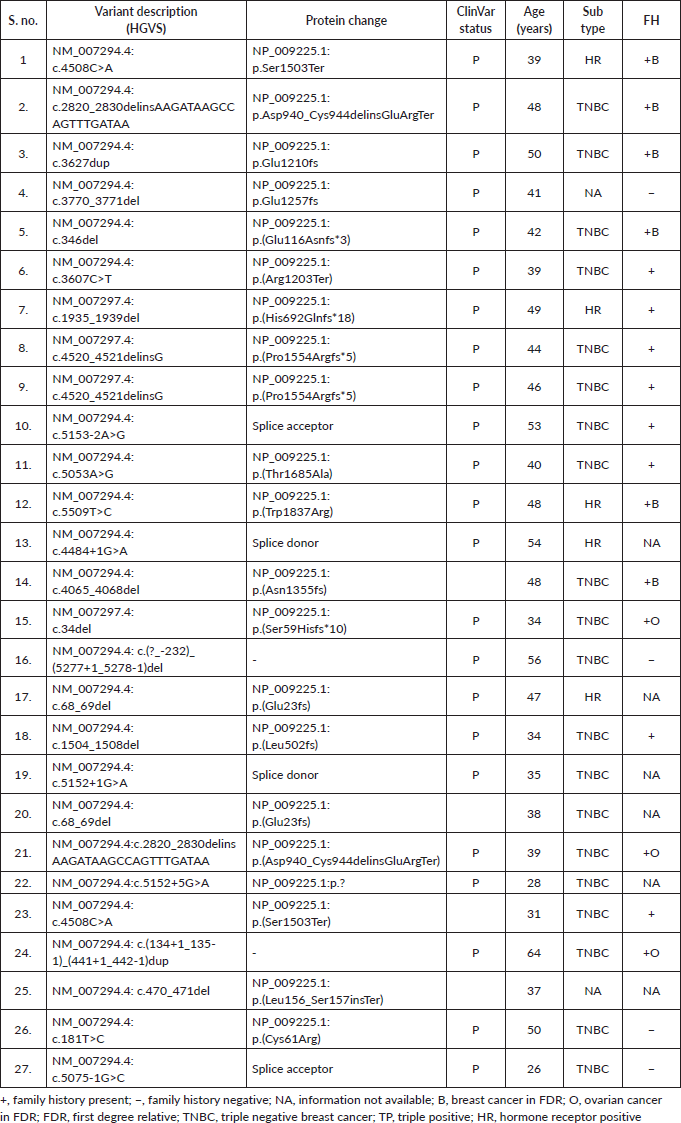
Supplementary Table S3b. P/LP variants detected in the BRCA2 gene.
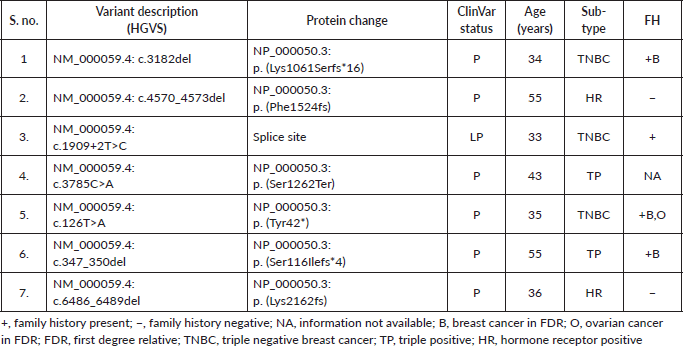
Supplementary Table S3c. P/LP variations in other genes.
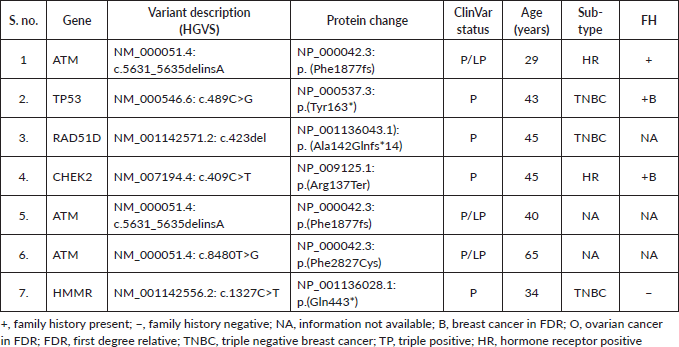
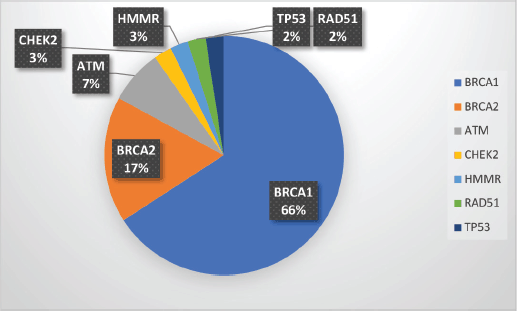
Supplementary Figure S1. Spectrum of P/LP mutations.
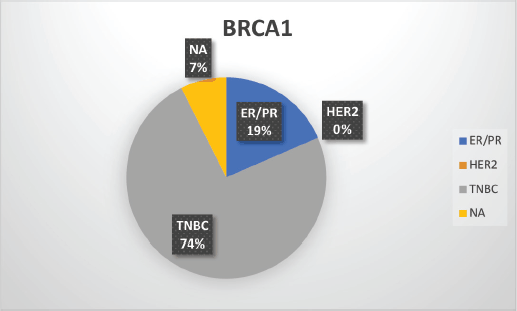
Supplementary Figure S2. Distribution of subtypes in BRCA1 patients.
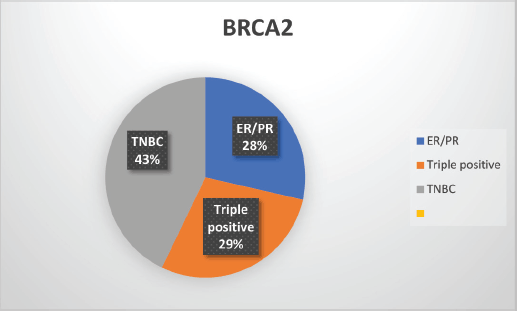
Supplementary Figure S3. Distribution of subtypes in BRCA2 patients.
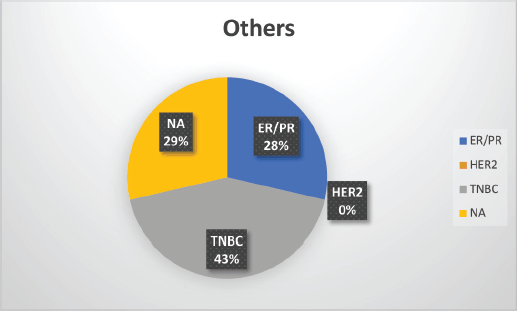
Supplementary Figure S4. Distribution of subtypes in non-BRCA mutation patients.